OVERVIEW OF BACTERIAL GENETIC MAPING By Shivaraj Guided

OVERVIEW OF BACTERIAL GENETIC MAPING By Shivaraj Guided by Prof. Girisha
Contents Introduction Conjugational analysis Gene libraries Restriction mapping &PFGE DNA sequence determination & Sanger sequencing 6. Pyrosequencing 7. Massively parallel sequencing 8. Conformation of gene by gene replacement 9. Metagenomics 10. Comparative genomics Microarrays 11. Real-time PCR 12. Analysis of gene expression using proteomics 1. 2. 3. 4. 5.
Introduction � Gene is a unit of inheritance, a piece of the genetic material that determines the inheritance of a particular characteristic, or group of characteristic. � Genes are carried by chromosomes in the cell nucleus and are arranged in a line along each chromosomes. � Every gene occupies a place, or locus, on the chromosome. � The word locus has become loosely interchangeable with the word GENE.
• The arrangement of Genes- arranged as a clusters of Genes are known as OPERONS. There are 2 types DNA sequences. � 1. EXONS- coding sequences. � 2. INTRONS- noncoding sequences. DNA Gene
� Genetic mapping 1. location 2. structure 3. and expression of the gene. � In the past carried out largely by using in vivo gene mapping. � Dramatic advancement in DNA sequencing. � Turning genomic sequence in to a meaningful information and how the gene expresses.
§ In bacteria the classical method of gene mapping depend on the production of recombinants by gene transfer, using conjugation, transformation and transduction. § These are largely replaced by methods based on in vitro gene technology. Chromosome
Conjugational analysis � Integration of F plasmid in to E. coli chromosome produce Hfr strain. � The transfer of whole chromosome would take 100 minute. � Each gene is assigned a position that corresponds to the time at which it transferred from the origin threonine locus (0 min ). � Transfer takes in clock wise direction. But it's quite rare for the complete chromosome to be transferred. � Prototrophic, auxotrophic , and minimal medium.
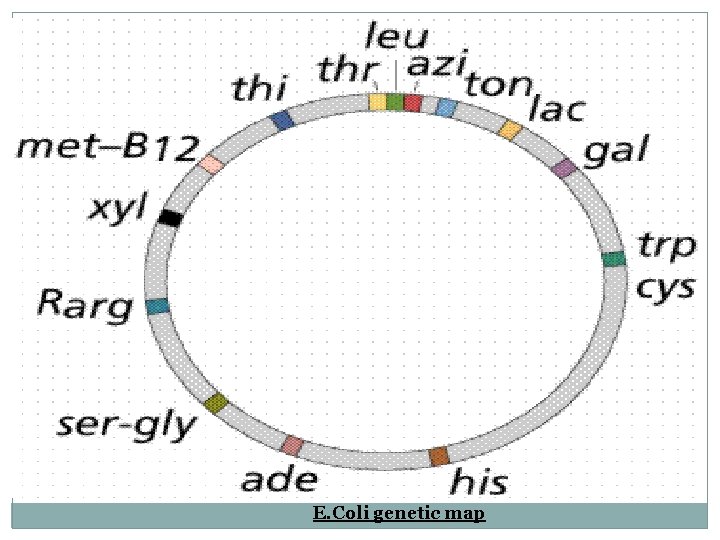
E. Coli genetic map
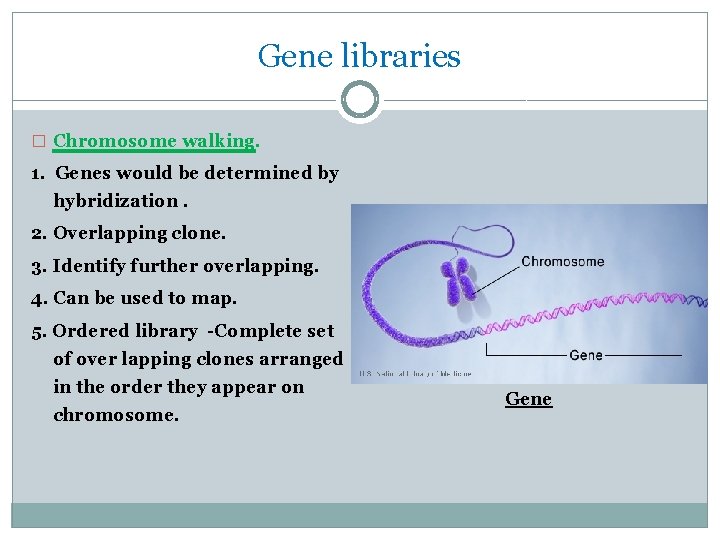
Gene libraries � Chromosome walking. 1. Genes would be determined by hybridization. 2. Overlapping clone. 3. Identify further overlapping. 4. Can be used to map. 5. Ordered library -Complete set of over lapping clones arranged in the order they appear on chromosome. Gene
Restriction mapping &PFGE � Use of restriction enzyme. � Use of enzyme which cuts in to long strand. � Like Not 1 & Pac 1. � PFGE. 1. Used to separate large DNA fragments. 2. Two direction separation. PFGE’s gel
3. Horizontal & vertical. 4. Normally the fragments move in same speed, but as we change the direction of the field they change they have to reorient themselves in the new direction, the longer the fragment, longer the time. � Separation based on size of the fragment. � This process is used to obtain fragments for metagenomics.
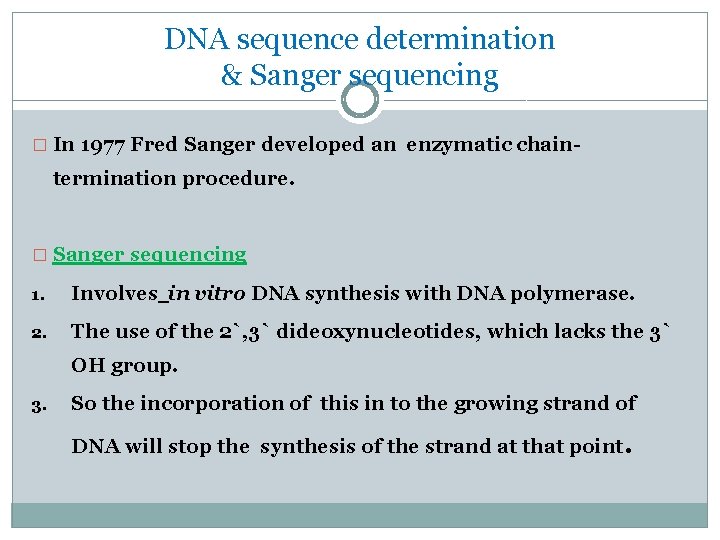
DNA sequence determination & Sanger sequencing � In 1977 Fred Sanger developed an enzymatic chain- termination procedure. � Sanger sequencing 1. Involves in vitro DNA synthesis with DNA polymerase. 2. The use of the 2`, 3` dideoxynucleotides, which lacks the 3` OH group. 3. So the incorporation of this in to the growing strand of DNA will stop the synthesis of the strand at that point.

4. The initiation of new the strand will start using a primer. 5. This reaction is supplemented with each of the four deoxynucleotide triphosphates and say dideoxy ATP. 6. Then at each point when an A residue should be added there is a chance of incorporating dd. A instead. 7. This will lead to a mixture of DNA chain each of which terminates at an A residue. 8. Running this on gel , will create a band which can be read.
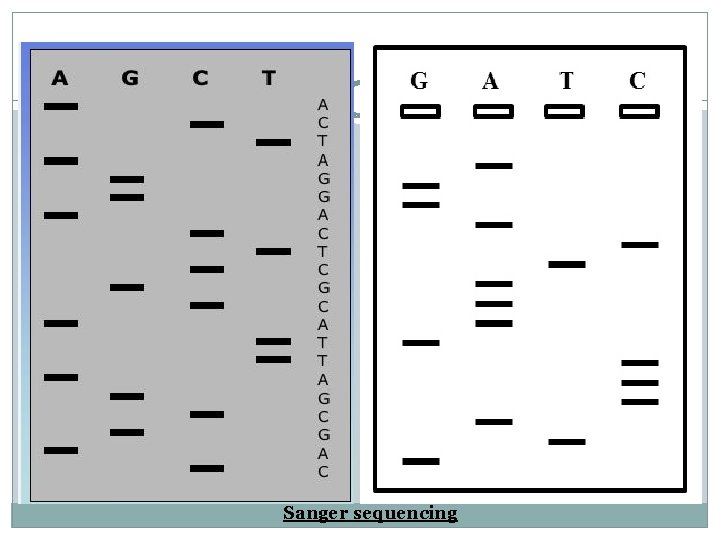
Sanger sequencing
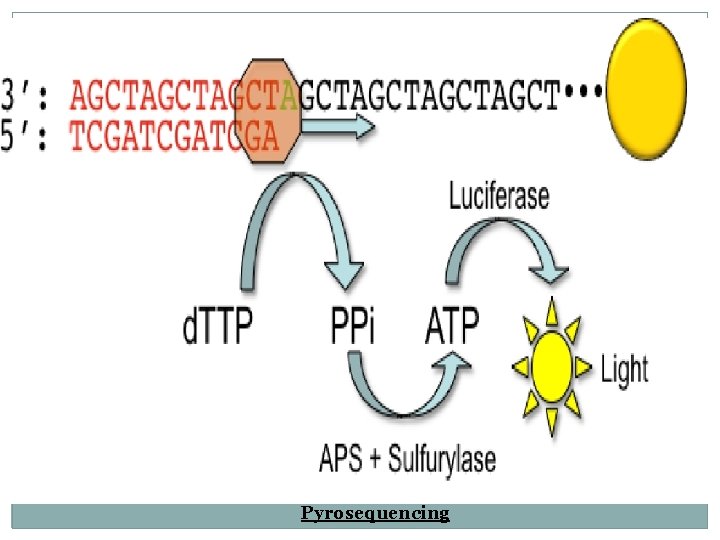
Pyrosequencing � Similar as of Sanger sequencing. � Relies on the detection of the release of pyrophosphate during nucleotide incorporation rather than the use of chain termination. � d. TTP is added for libration of PPi. � Pyrosequencing relies on the detection of these released pyrophosphate. � PPi in to detectable light.
Pyrosequencing
Pyrosequencing Kit

Illumina
Massively parallel sequencing � This type DNA sequencing is 100 times faster than pyrosequencing. � In this the DNA is broken up into small fragments and adaptor sequences are attached to the ends of the fragment. � Via the adaptor sequence the DNA is bound to microscopic beads (28µ in diameter ) so that only one genomic DNA fragment binds to each bead. � Now the fragments are needed to amplify, but its not possible to separate and amplify them. So they are encased in oil droplets which creates an individual tube that separates from each other.
� To perform the sequencing reaction, the DNA template carrying beads are loaded in to a specialized grid which contains 1. 6 million reaction vessels. � The small size of these ensures that only one bead can occupy one vessel and through this a grid with over 1 million separated DNA templates is generated. � The DNA sequence of each of the template is then determined simultaneously using the pyrosequencing chemistry.

Illumina

MPS
Conformation of gene by gene replacement � It is necessary to confirm that the gene which has been predicted has a specific function. � The gene is confirmed by inactivating the gene by mutation. � One of best way to find out the gene characters is to mutate the gene and find out the phenotype character. � The inactivation of gene is done in-vitro condition, by replacing the wild type gene by cloned one, this process is known as gene “knockout”.
Metagenomics � Metagenomics is the study of bacteria using genetic information from DNA, that is extracted directly from the environment with out culturing the bacteria. � Study of 16 s r. RAN sequence, which act as unique genetic barcode for each bacteria. Metagenomics 1. Isolation of DNA & RNA from a mixed population. 2. DNA fragmentation and construction of DNA library.
3. Sequencing at random or targeted fashion. 4. The clones are assembled in a continues pieces by matching up overlapping sequence. 5. The DNA or RNA sequence obtained are compared to database and noted by using bioinformatics. 6. When this method is coupled with pyrosequencing, it eliminates the need of cloning step. 7. It is one of the most powerful method to identify the diversity.
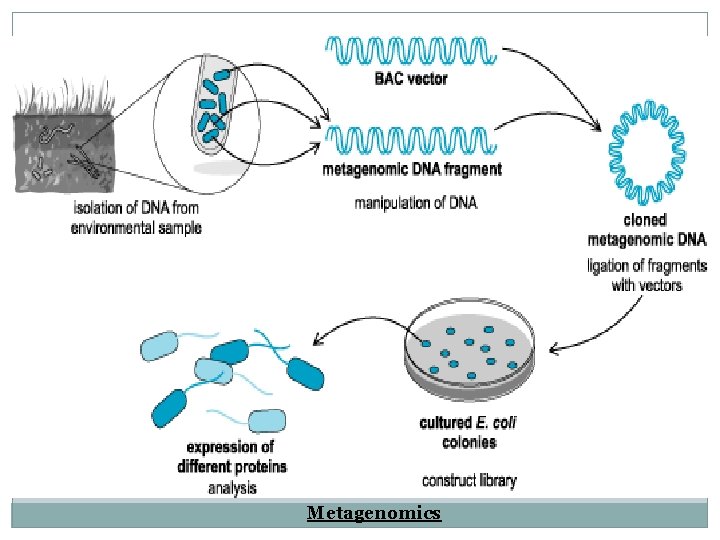
Metagenomics
Comparative genomics Microarrays � It is a comparative genomics where two genetic material of two different organisms are compared. � From a complete genome sequence it is possible to generate a set of DNA fragments that corresponds to a part of each gene in the genome, either by PCR or synthetic oligonuleotide. � This may seem impossible manually, for this reason robotic devices are used. � Tiny spots of each of these oligonuleotide is printed on to the surface of a glass slide in a grid-like arrangement again using robotics, Giving a precise positioning of each spot.
� To compare the genetic content of two bacteria (like of M. tuberculosis and B. calmette-Guerin) DNA from each organism is extracted and labeled with fluorescent dye. � The two DNA samples are mixed together and applied on the glass slide , so they will be hybridized to the array of spot. � Then the florescent is read by a machine that is able to distinguish the two dyes. � And the results are displaced as a pattern of dots. Red and green dots indicate binding of one or the other sample only, where as if both labeled DNA samples bind , the results will be shown as yellow dot.
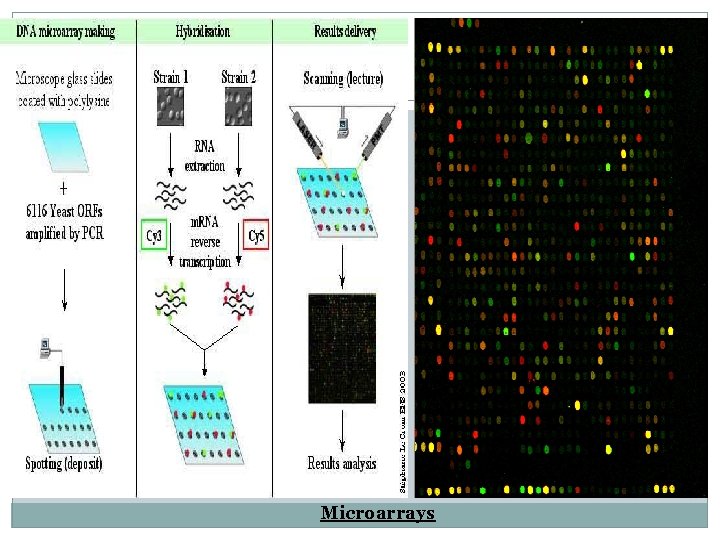
Microarrays
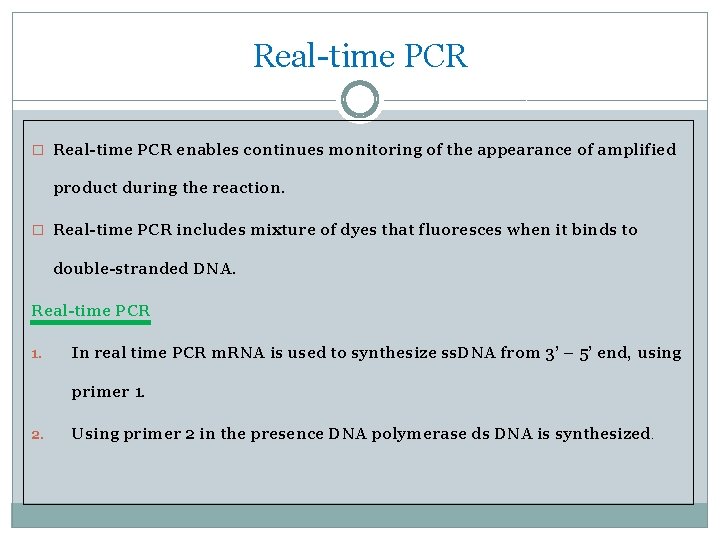
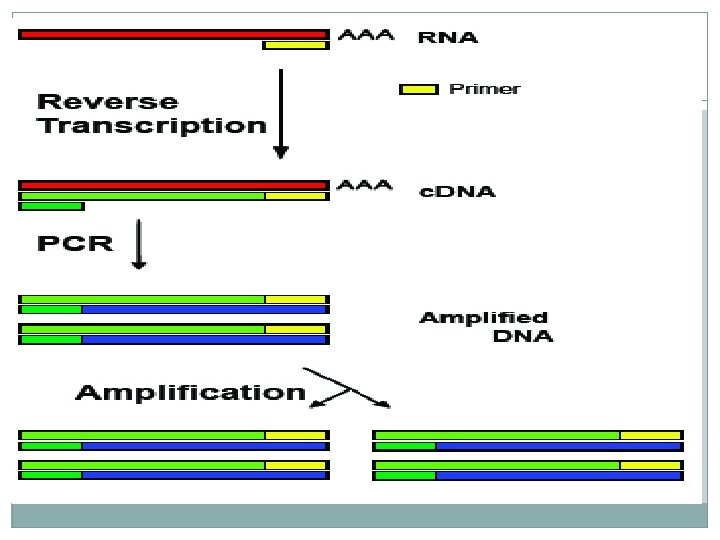
Real-time PCR � Real-time PCR enables continues monitoring of the appearance of amplified product during the reaction. � Real-time PCR includes mixture of dyes that fluoresces when it binds to double-stranded DNA. Real-time PCR 1. In real time PCR m. RNA is used to synthesize ss. DNA from 3’ – 5’ end, using primer 1. 2. Using primer 2 in the presence DNA polymerase ds DNA is synthesized .

3. Primer 2 & 3 synthesizes further strand on the newly synthesized strand, further amplifying the DNA strand. 4. During this process florescent dye is added which will illuminate when the specific nucleotide binds to the complementary strand. 5. This sensitive light is picked up by the machine, which will analyze the coding.
Real-time PCR

Analysis of gene expression using proteomics � For the synthesis of the protein transcription and translation process has to take place, this shows us that protein is descendent of the genetic material. � Each protein will have a unique amino acid sequencing which will be complementary to the DNA. � To analyze the genome the proteins have to be separated using 2 D PHAGE, where the separation will be according to charge and mass. � the separated spot will be removed and will be analyzed under NMR or MS, this will give the structure and sequencing of the protein.

2 D PHAGE

2 D PHAGE gel
Conclusion � The range of technique that we have today is redefining the way bacterial genetics and microbiology is practiced. � Remarkable technological advancement mean that it is now possible to obtain the genome sequence of a bacterium in less than a day. � As a result of this, today's scientists no longer spend years characterizing a single gene or protein in isolation, but are now able to characterize hundreds of different biomolecules simultaneously. � How ever to make the most of this information it has to be related back to the behavior of the cell as a whole. � The unification of these biological data will be the major challenge in the years ahead.

Reference � Larry snyder & wendy champness, molecular genetics of bacteria, 2 nd ed 2006, American society for microbiology, USA. � Madigan, Martinko, Stahl, Clark, Biology of microorganisms, 13 th ed, 2012, Pearson, USA. � Jeremy W dale, Simon F park, molecular genetics of bacteria, 5 th ed , 2016, Wiley, USA.

- Slides: 39