Overview of ALS Genetics Parts of whats known

![Human Genome Basics • Distinctive markers exist all across the genome. o [Large] RFLP, Human Genome Basics • Distinctive markers exist all across the genome. o [Large] RFLP,](https://slidetodoc.com/presentation_image/784f4def4d5eeee51233cdbab70f57ef/image-3.jpg)



![Superoxide Dismutase-1 SOD 1 [ALS 1] • Originally found using Classical linkage analysis and Superoxide Dismutase-1 SOD 1 [ALS 1] • Originally found using Classical linkage analysis and](https://slidetodoc.com/presentation_image/784f4def4d5eeee51233cdbab70f57ef/image-9.jpg)
![TAR DNA Binding Protein TARDBP (TDP-43) [ALS 10] • [Candidate gene approach] • In TAR DNA Binding Protein TARDBP (TDP-43) [ALS 10] • [Candidate gene approach] • In](https://slidetodoc.com/presentation_image/784f4def4d5eeee51233cdbab70f57ef/image-10.jpg)
![Fused in Sarcoma FUS [ALS 6] • [Candidate gene approach and locus approaches] (2009) Fused in Sarcoma FUS [ALS 6] • [Candidate gene approach and locus approaches] (2009)](https://slidetodoc.com/presentation_image/784f4def4d5eeee51233cdbab70f57ef/image-11.jpg)
![Chromosome 9 open reading frame C 9 orf 72 [ALS-FTD] • [GWAS and locus Chromosome 9 open reading frame C 9 orf 72 [ALS-FTD] • [GWAS and locus](https://slidetodoc.com/presentation_image/784f4def4d5eeee51233cdbab70f57ef/image-12.jpg)




- Slides: 21

Overview of ALS Genetics Parts of what’s known and a glimpse of what’s next… Patrick Dion, Ph. D. Neurology and Neurosurgery Mc. Gill University

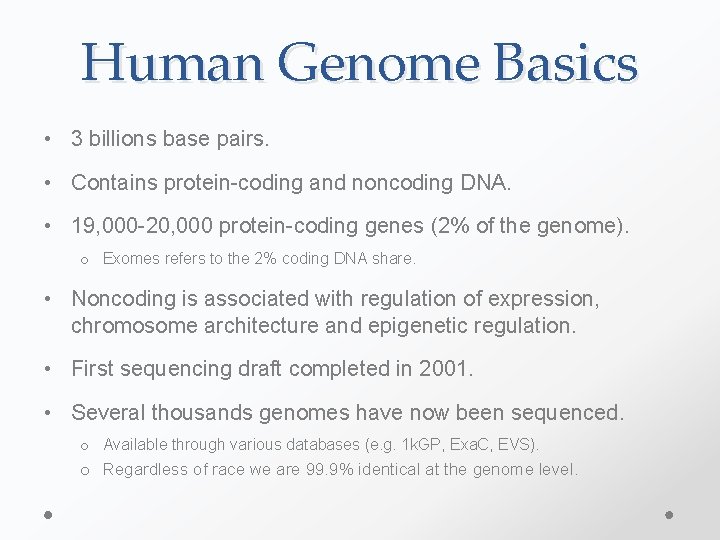
Human Genome Basics • 3 billions base pairs. • Contains protein-coding and noncoding DNA. • 19, 000 -20, 000 protein-coding genes (2% of the genome). o Exomes refers to the 2% coding DNA share. • Noncoding is associated with regulation of expression, chromosome architecture and epigenetic regulation. • First sequencing draft completed in 2001. • Several thousands genomes have now been sequenced. o Available through various databases (e. g. 1 k. GP, Exa. C, EVS). o Regardless of race we are 99. 9% identical at the genome level.
![Human Genome Basics Distinctive markers exist all across the genome o Large RFLP Human Genome Basics • Distinctive markers exist all across the genome. o [Large] RFLP,](https://slidetodoc.com/presentation_image/784f4def4d5eeee51233cdbab70f57ef/image-3.jpg)
Human Genome Basics • Distinctive markers exist all across the genome. o [Large] RFLP, tandem repeat, Copy Number Variants (CNVs) o [Small] Single nucleotide polymorphisms (SNPs). • SNPs are common variants found in >1% of the population at 1, 000 bp across. o When in coding regions SNPs can be synonymous or nonsynonymous. • Distinction between SNPs and single nucleotide variants (SNVs).

Discovery Approaches • Classical linkage analysis • Candidate genes association • Genome Wide Association Study (GWAS) o Use of common variants to identify regions shared by affected. • Whole Exome Sequencing (WES) o Seeks to filter rare coding variants to identify deleterious ones. o No need for controls except when population specific. • Whole Genome Sequencing (WGS) o Seeks to filter all variants (coding and not coding) • WES and WGS data can also be used to conduct association studies using common variants and rare ones (e. g. Minor Allele Frequency < 1%)

Variants filtering of WES and WGS

FALS and SALS • Overall ALS is the most common rare disease (2/100, 00). • FALS (familial) o 5 -10% of cases of ALS o Primarily autosomal dominant (AD) segregation of definite, probable or possible individuals. • SALS (sporadic) o No family history • Clinically indistinguishable o Except age of onset and sex distribution • This classification should not overshadow the fact that FALS and SALS DO share common genetic causes. o Moreover environmental/stochastic factors can affect genetically susceptible individuals.

Just how many ALS genes are there? • http: //alsod. iop. kcl. ac. uk/home. aspx o Updated once a year. • Between the discovery of the first causative gene (SOD 1) in 1993 and now, 126 genes were “linked” to ALS. • [+] Classify the genes according to phenotype, geographical distribution and method of identification. • [-] Includes > 50% of genes for which associations are either weak and/or were never replicated in independent studies. • Nonetheless a valuable and objective online to use assessment tool to consult about the causal nature of specific variants. • The risk linked to ALS genes can be classified according to multiple factors.

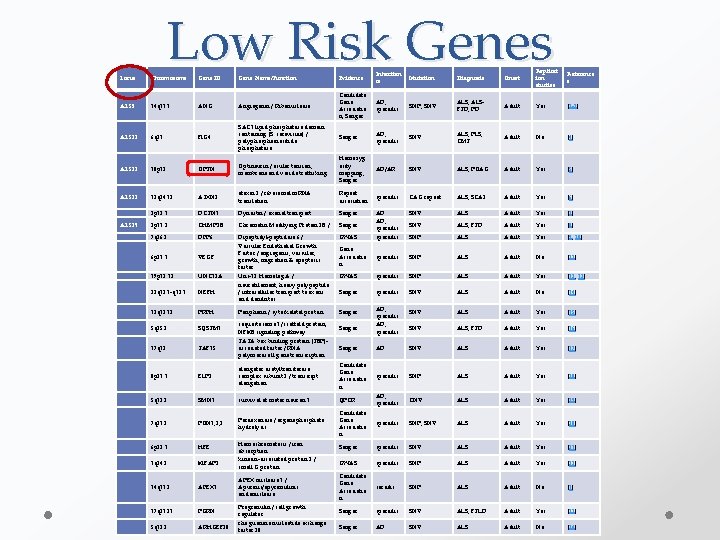
High Risk Genes Locus Chromoso me Gene ID Protein/Function Evidence Inheritan ce Mutatio n Diagnosis Onset Replicati on studies Reference s ALS 1 21 q 22. 11 SOD 1 Cu/Zn superoxide dismutase 1, soluble / Oxidative stress Linkage, Sanger AD, AR SNV ALS, PMA Adult Yes [1] ALS 2 2 q 33. 2 ALS 2 Alsin / Rho guanine nucleotide exchange factors Linkage, Sanger AR SNV ALS, PLS, HSP Juvenile Yes [2, 3] ALS 4 9 q 34. 13 SETX Linkage, Sanger AD SNV ALS, AOA 2 Juvenile Yes [4] ALS 5 15 q 21. 1 SPG 11 Linkage, Sanger AR SNV ALS, HSP Juvenile Yes [5, 6] ALS 6 16 p 11. 2 FUS Linkage, Sanger AD SNV ALS, ALSFTD Adult Yes [7, 8] ALS 7 20 p 13 Unknown Linkage AD - ALS Adult No [9] ALS 8 20 q 13. 33 VAPB Linkage, Sanger AD SNV ALS, SMA Adult Yes [10, 11] ALS 10 1 p 36. 22 TARDBP TAR DNA binding protein 43 / transcriptional repressor, splicing regulation Linkage, Sanger AD SNV ALS, ALSFTD Adult Yes [12, 13] ALS 14 9 p 13. 3 VCP valosin-containing protein / ATP-binding protein, vesicle transport and fusion WES, linkage, Sanger AD SNV ALS, ALSFTD, IBMPFD Adult Yes [14] ALS 15 Xp 11. 21 UBQLN 2 ubiquilin 2 / ubiquitination, degradation Linkage, Sanger X-linked SNV ALS, ALSFTD Juvenile, Adulte Yes [15] sigma non-opioid intracellular receptor 1 / endoplasmic reticulum chaperone Homozygosity mapping, Sanger AR SNV ALS, FTD Juvenile No [15] Linkage, Sanger AD SNV ALS Adult No [16] Linkage, WES, Sanger AD SNV ALS Adult Yes [17] Senataxin / RNA/DNA Helicase Spatacsin / transmembrane protein Fused in Sarcoma / RNAbinding protein, DNA repair, exon splicing Unknown Vesicle-associated membrane proteinassociated protein B / Vesicular trafficking ALS 16 9 p 13. 3 SIGMAR 1 12 q 24. 11 DAO ALS 17 17 p 13. 2 PFN 1 ALS 20 7 p 15. 2/12 q 13. 3 hn. RNPA 2 B 1/A 1 Heterogenous nuclear ribonucleoprotein / m. RNA processing, metabolism & transport Linkage, WES, Sanger AD SNV ALS, IBMPFD Adult No [18] ALSFTD 1 9 q 21 -q 22 Unknown Linkage AD SNV ALS, ALSFTD, FTD Adult No [19] ALSFTD 2 9 p 21. 2 C 9 ORF 72 chromosome 9 open reading frame 72 / Unknown Linkage, GWAS, RP-PCR, Southern Blot AD, sporadic 1 G 4 C 2 repeat ALS, ALSFTD, FTD Adult Yes [20 -28] D-amino-acid oxidase / unknown profilin 1 / actin binding protein, actin polymerization
![Superoxide Dismutase1 SOD 1 ALS 1 Originally found using Classical linkage analysis and Superoxide Dismutase-1 SOD 1 [ALS 1] • Originally found using Classical linkage analysis and](https://slidetodoc.com/presentation_image/784f4def4d5eeee51233cdbab70f57ef/image-9.jpg)
Superoxide Dismutase-1 SOD 1 [ALS 1] • Originally found using Classical linkage analysis and FALS. • Accounts for ~20% of FALS forms (2% of SALS). • >160 mutations identified all over SOD 1. o All dominant except for D 90 A and D 96 N, recessive in some cases o Some mutations affect survival time: • A 4 V rapid progression D 90 A slow progression o Some affect disease onset: • G 37 R with early onset o Most mutations trigger a shift of the folding equilibrium toward poorly structured SOD monomers. • A great number of mechanisms are proposed to be involved, however, distinguishing cause from effect and identifying the critical processes remains challenging
![TAR DNA Binding Protein TARDBP TDP43 ALS 10 Candidate gene approach In TAR DNA Binding Protein TARDBP (TDP-43) [ALS 10] • [Candidate gene approach] • In](https://slidetodoc.com/presentation_image/784f4def4d5eeee51233cdbab70f57ef/image-10.jpg)
TAR DNA Binding Protein TARDBP (TDP-43) [ALS 10] • [Candidate gene approach] • In 2008 the gene was screened for mutations as its product was a prominent product of ubiquitinated cytoplasmic inclusions in the CNS tissues of FTD and ALS. • Accounts for ~4% of FALS forms (<1 % of SALS). • > 47 Missense and one truncating variants. o All variants are dominants. • Pathogenic variants are mostly in the C-terminus which is involved in RNA binding and splicing.
![Fused in Sarcoma FUS ALS 6 Candidate gene approach and locus approaches 2009 Fused in Sarcoma FUS [ALS 6] • [Candidate gene approach and locus approaches] (2009)](https://slidetodoc.com/presentation_image/784f4def4d5eeee51233cdbab70f57ef/image-11.jpg)
Fused in Sarcoma FUS [ALS 6] • [Candidate gene approach and locus approaches] (2009) • Accounts for ~4% of FALS forms (<1 % of SALS). o Autosomal dominant and recessive. o Age of onset younger (< 40 yrs with cases during teens). o Faster progression than TARDBP and SOD 1 cases • > 50 Missense and one truncating variants. o All variants are dominants. • Pathogenic variants are mostly in the C-terminus.
![Chromosome 9 open reading frame C 9 orf 72 ALSFTD GWAS and locus Chromosome 9 open reading frame C 9 orf 72 [ALS-FTD] • [GWAS and locus](https://slidetodoc.com/presentation_image/784f4def4d5eeee51233cdbab70f57ef/image-12.jpg)
Chromosome 9 open reading frame C 9 orf 72 [ALS-FTD] • [GWAS and locus approaches] (2011) • Accounts for ~40% of European descent familial ALS-FTD cases ( 10 % of Asians) and 7% of SALS. • Large intronic repeat expansion (GGGGCC or G 4 C 2). o >30 repeats pathogenic, 15 -30 not very pathogenic but recently observed to be with ATXN 2 intermediate CAG expansion. o Both gain and loss of functions are under consideration. o Multiple pathogenic avenues. . • RNA foci and sequestration of RNA binding components. • Non-ATG (RAN) translation of repeat derived dipeptide accumulation in the CNS (GR > PR > GA > AP > GP). • Dysregulation of its potential DENN Rab-GEFs activity on membrane trafficking. • Disrupts of nucleocytoplasmic transport of m. RNA.

Low Risk Genes Locus Chromosome Gene ID Gene Name/Function Evidence Inheritan ce Mutation Diagnosis Onset Replicat ion studies Reference s ALS 9 14 q 11. 1 ANG Angiogenin / Ribonuclease Candidate Gene Associatio n, Sanger AD, sporadic SNP, SNV ALS, ALSFTD, PD Adult Yes [1 -3] ALS 11 6 q 21 FIG 4 SAC 1 lipid phosphatase domain containing (S. cerevisiae) / polyphosphoinositide phosphatase Sanger AD, sporadic SNV ALS, PLS, CMT Adult No [4] ALS 12 10 p 13 OPTN Optineurin / ocular tension, membrane and vesicle trafficking Homozyg osity mapping, Sanger AD/AR SNV ALS, POAG Adult Yes [5] ALS 13 12 q 24. 12 ATXN 2 ataxin 2 / ribosomal m. RNA translation Repeat association sporadic CAG repeat ALS, SCA 2 Adult Yes [6] 2 p 13. 1 DCTN 1 Dynactin / axonal transport Sanger ALS Adult Yes [7] 3 p 11. 2 CHMP 2 B Chromatin Modifying Protein 2 B / Sanger SNV ALS, FTD Adult Yes [8] 7 q 36. 2 DPP 6 Dipeptidyl-peptidase 6 / GWAS AD AD, sporadic SNV ALS 17 SNP ALS Adult Yes [9, 10] Vascular Endothelial Growth Factor / angiogenic, vascular, growth, migration & apoptosis factor Gene Associatio n sporadic SNP ALS Adult No [11] Unc-13 Homolog A / neurofilament, heavy polypeptide / intracellular transport to axons and dendrites GWAS sporadic SNP ALS Adult Yes [12, 13] Sanger sporadic SNV ALS Adult No [14] SNV ALS Adult Yes [15] SNV ALS, FTD Adult Yes [16] 6 p 21. 1 VEGF 19 p 13. 12 UNC 13 A 22 q 12. 1 -q 13. 1 NEFH 12 q 13. 12 PRPH Peripherin / cytoskeletal protein Sanger SQSTM 1 sequestosome 1 /scaffold protein, NFKB signaling pathway Sanger AD SNV ALS Adult Yes [17] 5 q 35. 3 AD, sporadic 17 q 12 TAF 15 TATA box binding protein (TBP)associated factor / RNA polymerase II gene transcription 8 p 21. 1 ELP 3 elongator acetyltransferase complex subunit 3 / transcript elongation Candidate Gene Associatio n sporadic SNP ALS Adult Yes [18] 5 q 13. 2 SMN 1 survival of motor neuron 1 QPCR AD, sporadic CNV ALS Adult Yes [19] Paraoxonase / organophosphate hydrolysis Candidate Gene Associatio n sporadic SNP, SNV ALS Adult Yes [20] Sanger sporadic SNV ALS Adult Yes [21] GWAS sporadic SNP ALS Adult Yes [22] Candidate Gene Associatio n soradic SNP ALS Adult No [1] Sanger sporadic SNV ALS, FTLD Adult Yes [23] Sanger AD SNV ALS Adult No [24] 7 q 21. 3 PON 1, 2, 3 6 p 22. 1 HFE 1 q 24. 2 KIFAP 3 14 q 11. 2 APEX 1 17 q 21. 31 PGRN 5 q 13. 2 ARHGEF 28 Hemochromatosis / iron absorption kinesin-associated protein 3 / small G protein APEX nuclease 1 / Apurinic/apyrimidinic endonuclease Progranulin / cell growth regulator rho guanine nucleotide exchange factor 28

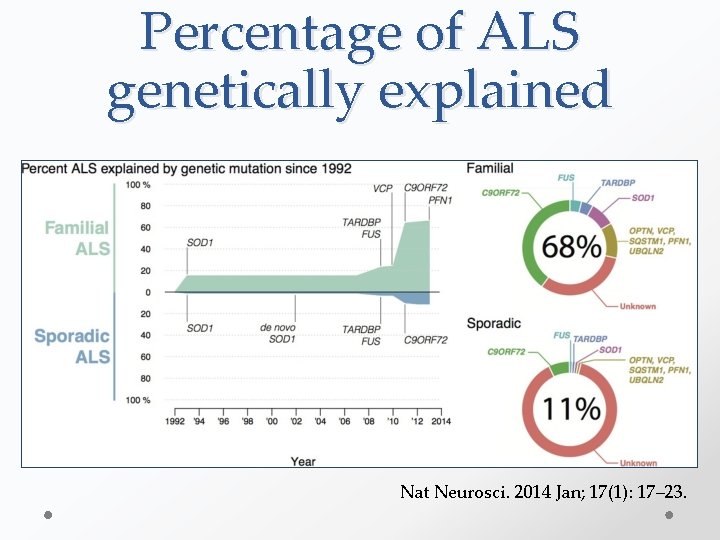
Percentage of ALS genetically explained Nat Neurosci. 2014 Jan; 17(1): 17– 23.

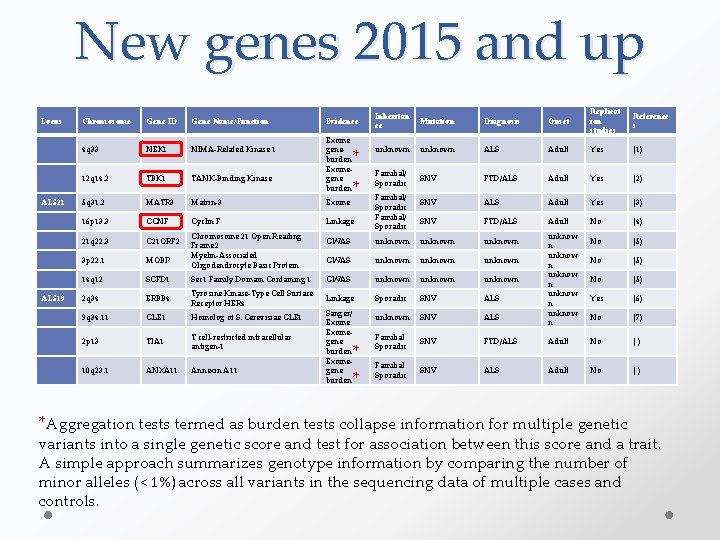
New genes 2015 and up Locus Chromosome Gene ID Gene Name/Function 4 q 33 NEK 1 NIMA-Related Kinase 1 12 q 14. 2 TBK 1 TANK-Binding Kinase ALS 21 5 q 31. 2 MATR 3 Matrin-3 Exome 16 p 13. 3 CCNF Cyclin F Linkage 21 q 22. 3 C 21 ORF 2 3 p 22. 1 MOBP 14 q 12 SCFD 1 ALS 19 2 q 34 ERBB 4 Chromosome 21 Open Reading Frame 2 Myelin-Associated Oligodendrocyte Basic Protein Evidence Exome geneburden Exomegene burden * * Inheritan ce Mutation Diagnosis Onset Replicat ion studies Reference s unknown ALS Adult Yes (1) Familial/ Sporadic SNV FTD/ALS Adult Yes (2) SNV ALS Adult Yes (3) SNV FTD/ALS Adult No (4) No (5) Yes (6) No (7) Familial/ Sporadic unknow n unknow n GWAS unknown unknown Sec 1 Family Domain Containing 1 GWAS unknown Tyrosine Kinase-Type Cell Surface Receptor HER 4 Linkage Sporadic SNV ALS unknown SNV ALS Familial Sporadic SNV FTD/ALS Adult No ( ) Familial Sporadic SNV ALS Adult No ( ) 9 q 34. 11 GLE 1 Homolog of S. Cerevisiae GLE 1 2 p 13 TIA 1 T cell-restricted intracellular antigen-1 10 q 23. 1 ANXA 11 Annexin A 11 Sanger/ Exomegene burden * * *Aggregation tests termed as burden tests collapse information for multiple genetic variants into a single genetic score and test for association between this score and a trait. A simple approach summarizes genotype information by comparing the number of minor alleles (˂ 1%) across all variants in the sequencing data of multiple cases and controls.

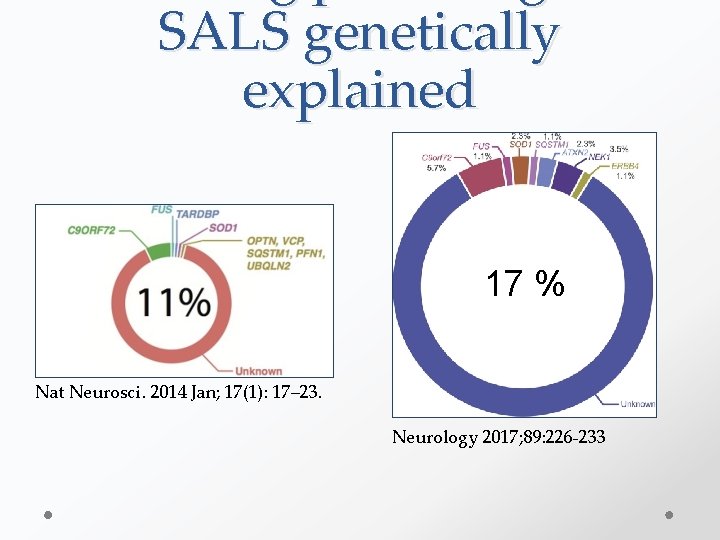
SALS genetically explained 17 % Nat Neurosci. 2014 Jan; 17(1): 17– 23. Neurology 2017; 89: 226 -233

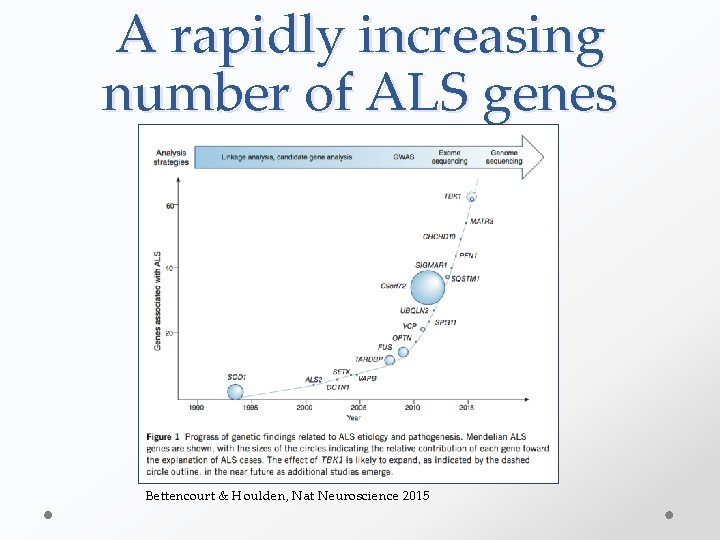
A rapidly increasing number of ALS genes Bettencourt & Houlden, Nat Neuroscience 2015

Rate of ALS gene Discovery >100 ALS genes? C 21 ORF 2 MOBP SCFD 1 CCNF 2017 Rate of new gene discovery has reached a plateau despite increased sequencing efforts UNC 13 A TIA 1 ANXA 11

Rate of gene discovery • “Plateau” Stage o Several candidate genes without strong evidence o Functional relevance unknown • Limit reached for current technologies o Require larger cohorts to detect small effect sizes • Deep phenotyping and extreme phenotypes • New directions: • • Large consortia (Min. E) Whole genome sequencing Rare and small structural variants Epigenetics

Other themes tested for ALS Genetics • Oligogenicity o Possibility that ALS is caused by two (or more) variants concurrently that would not independently cause disease. o Studies have observed patients with multiple mutations in “causal” genes. • e. g. C 9 orf 72 expansion with an OPTN mutation of unknown pathogenicity o Does severity of disease increase with more ALS mutations? • De novo mutations o Mutations can occur in germinal cells, not present in either parent • Sporadic ALS patients have no family history, but could pass new mutations to children? o De novo is not commonly observed in genetic studies, not a common cause of ALS

Thank you! patrick. a. dion@mcgill. ca