Outline Sequence alignment Algorithm Parallel Identification and mining









- Slides: 63


Outline • Sequence alignment – Algorithm – Parallel • Identification and mining – micro. RNA – machine learning related works • Function prediction – mi. RNA disease relationship – crops yield related genes Tianjin University

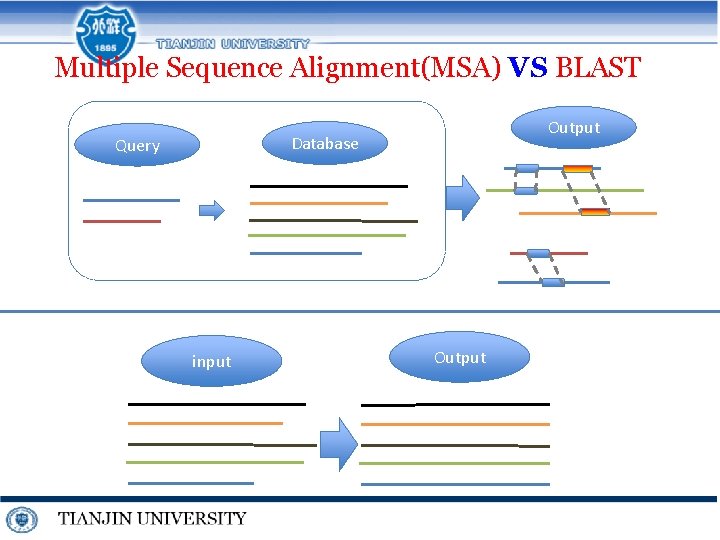
Multiple Sequence Alignment(MSA) VS BLAST Output Database Query input Output

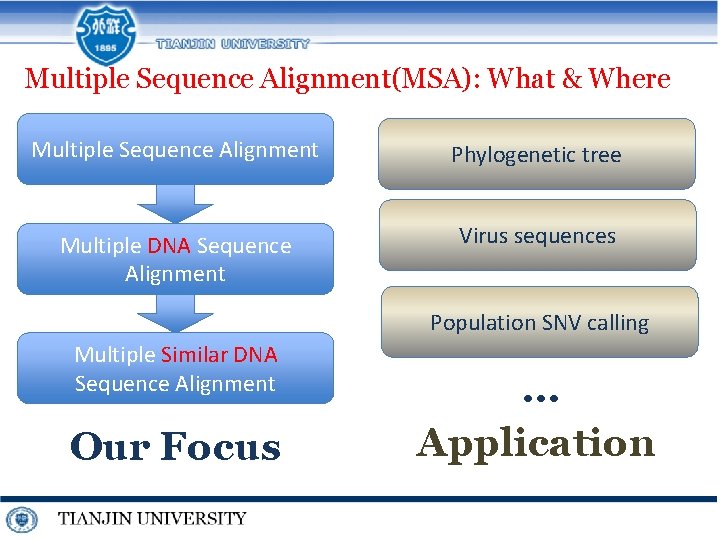
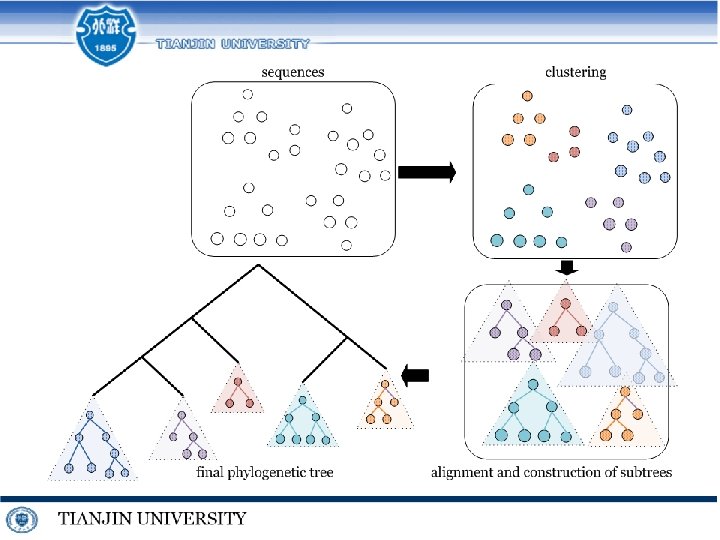
Multiple Sequence Alignment(MSA): What & Where Multiple Sequence Alignment Phylogenetic tree Multiple DNA Sequence Alignment Virus sequences Population SNV calling Multiple Similar DNA Sequence Alignment Our Focus … Application

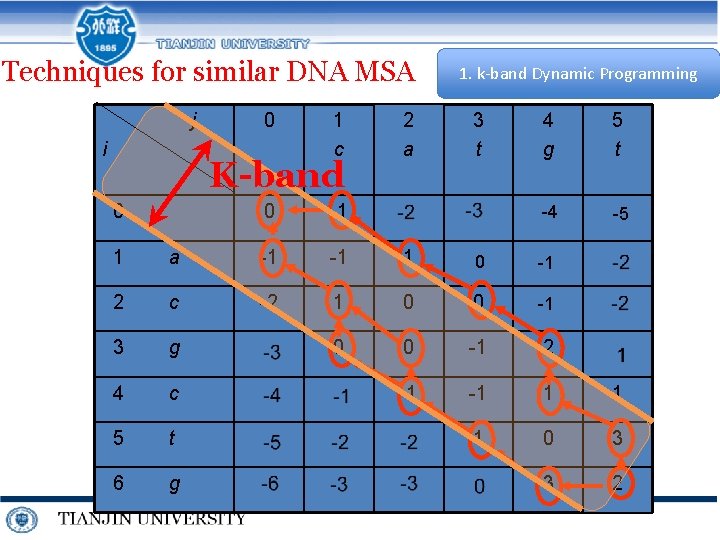
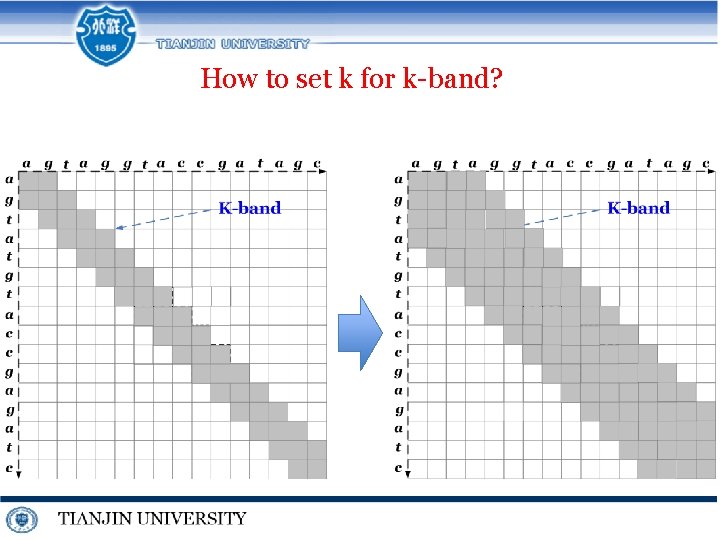
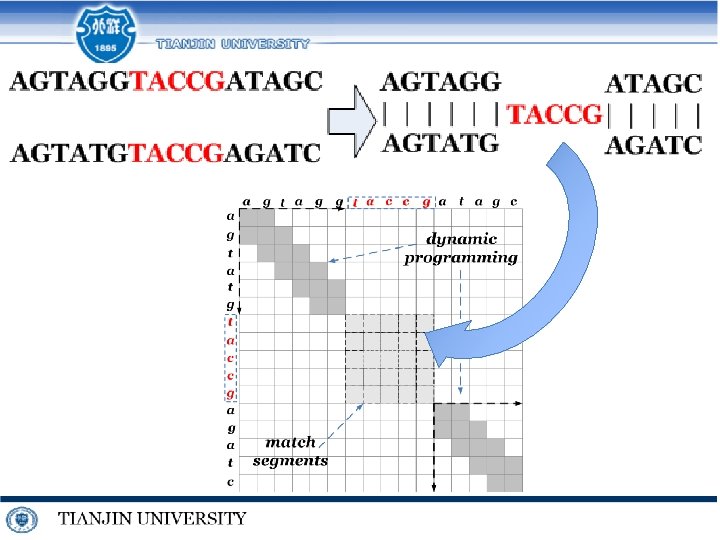
Techniques for similar DNA MSA j i 0 1 c K-band 0 0 -1 2 a 1. k-band Dynamic Programming 3 t 4 g 5 t -4 -5 1 a -1 -1 1 0 -1 2 c -2 1 0 0 -1 3 g 0 0 -1 2 4 c -1 -1 1 1 5 t 1 0 3 6 g 3 2

How to set k for k-band?


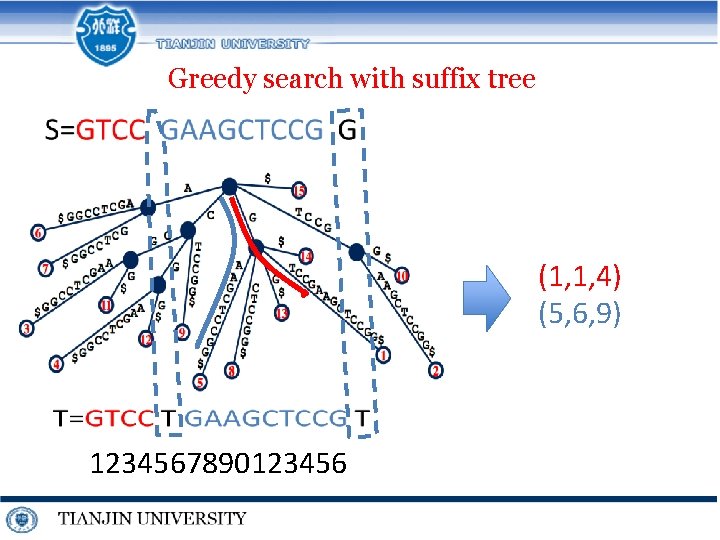
Greedy search with suffix tree S=GTCCGAAGCTCCGG (1, 1, 4) (5, 6, 9) T=GTCCTGAAGCTCCGT 1234567890123456

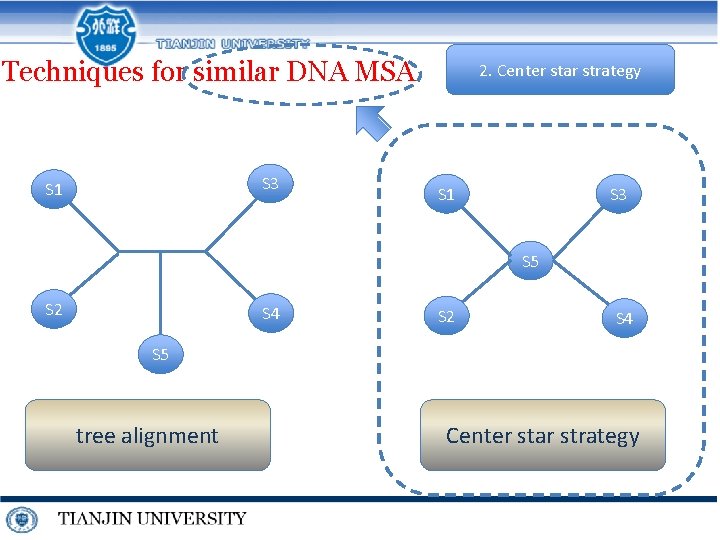
Techniques for similar DNA MSA S 3 S 1 2. Center star strategy S 1 S 3 S 5 S 2 S 4 S 5 tree alignment Center star strategy

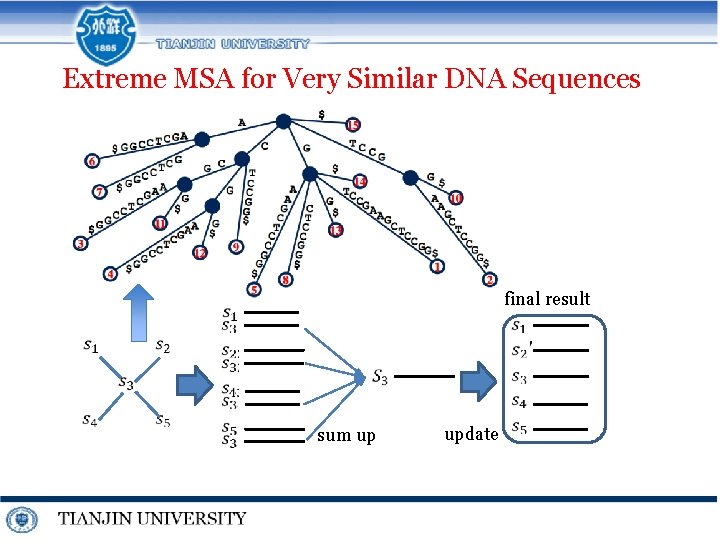
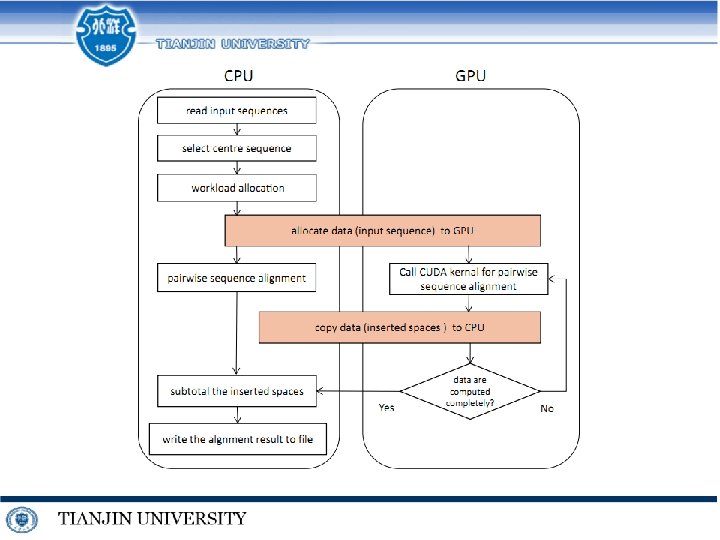
Extreme MSA for Very Similar DNA Sequences final result sum up update

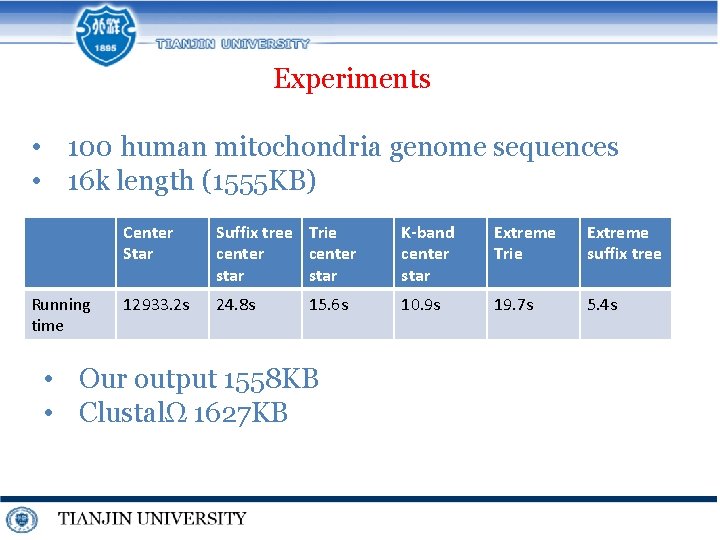
Experiments • 100 human mitochondria genome sequences • 16 k length (1555 KB) Running time Center Star Suffix tree Trie center star K-band center star Extreme Trie Extreme suffix tree 12933. 2 s 24. 8 s 10. 9 s 19. 7 s 5. 4 s 15. 6 s • Our output 1558 KB • ClustalΩ 1627 KB

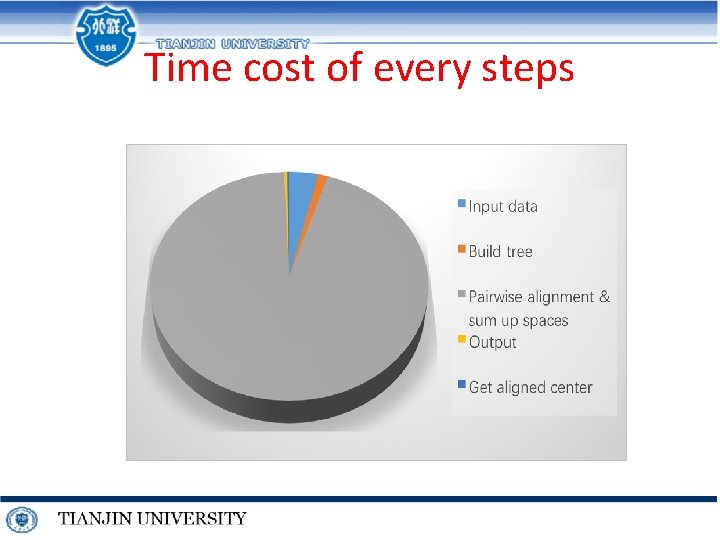
Time cost of every steps

Outline • Sequence alignment – Algorithm – Parallel • Identification and mining – micro. RNA – machine learning related works • Function prediction – mi. RNA disease relationship – crops yield related genes

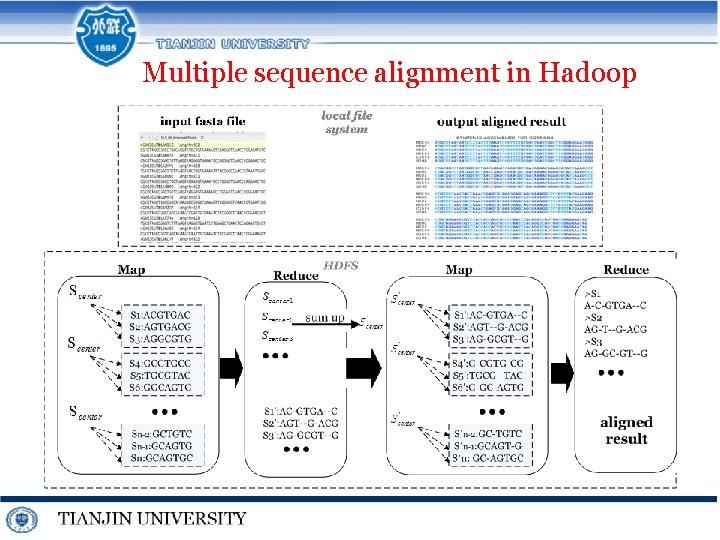
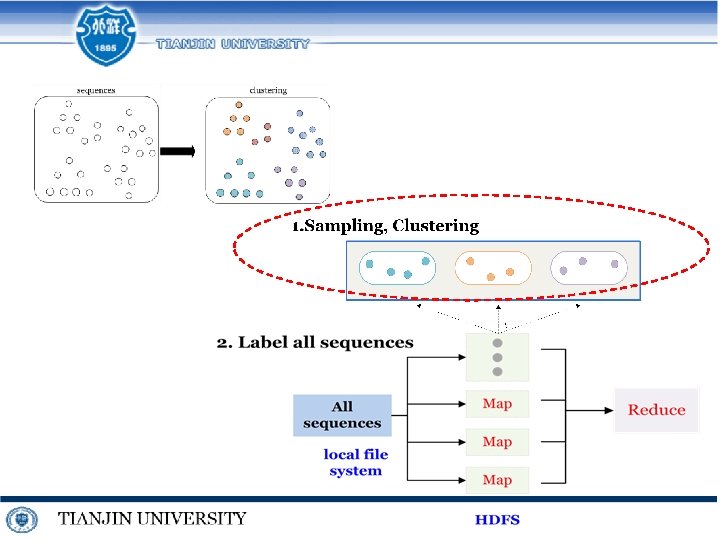
Multiple sequence alignment in Hadoop

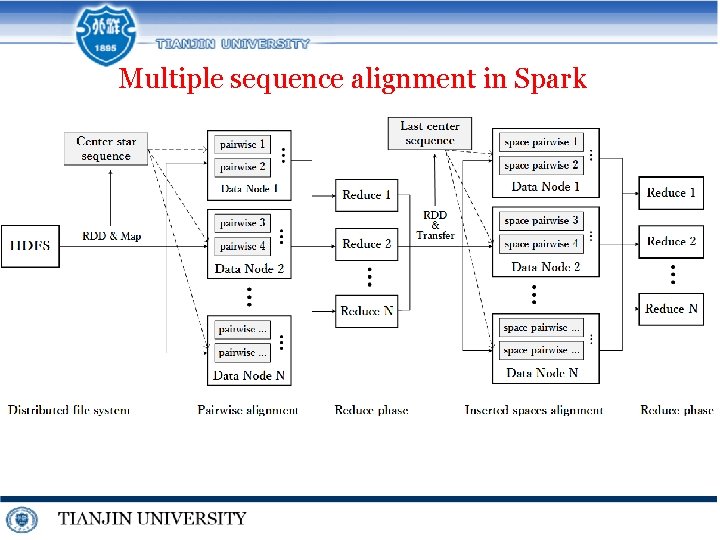
Multiple sequence alignment in Spark

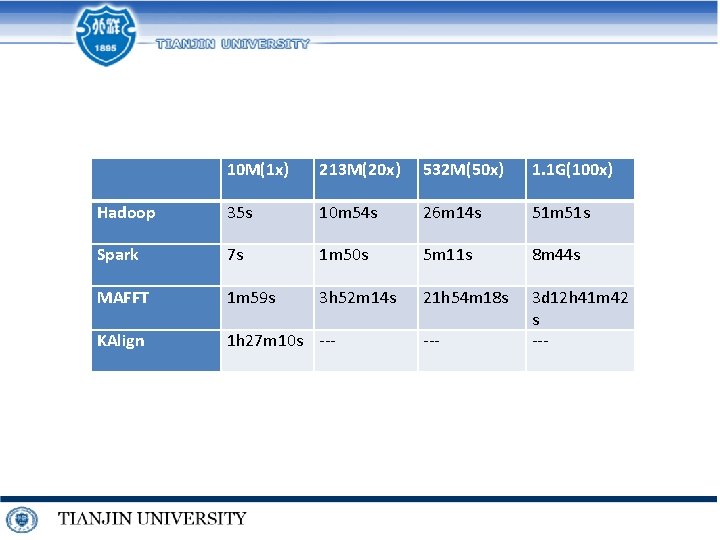
10 M(1 x) 213 M(20 x) 532 M(50 x) 1. 1 G(100 x) Hadoop 35 s 10 m 54 s 26 m 14 s 51 m 51 s Spark 7 s 1 m 50 s 5 m 11 s 8 m 44 s MAFFT 1 m 59 s 3 h 52 m 14 s 21 h 54 m 18 s KAlign 1 h 27 m 10 s --- 3 d 12 h 41 m 42 s ---




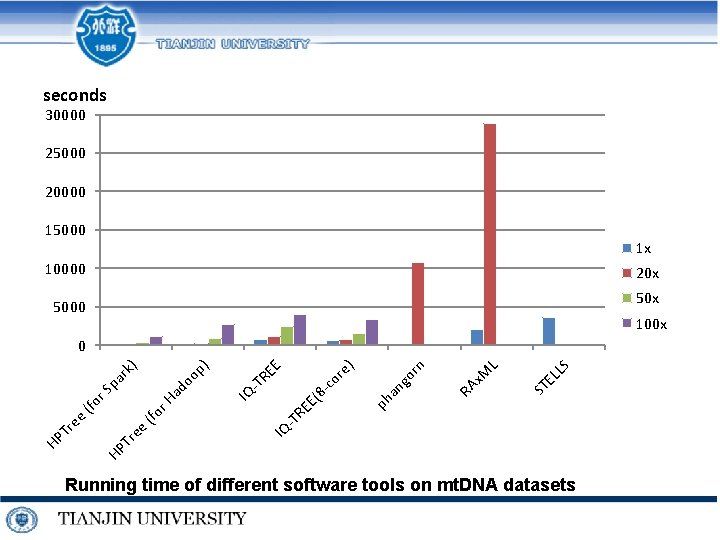
seconds 30000 25000 20000 15000 1 x 10000 20 x 5000 100 x LS EL ST L x. M RA rn go an ph IQ -T RE E( 8 c or e) E -T RE IQ oo r. H (fo Tr ee HP HP Tr ee (fo r. S ad pa rk ) p) 0 Running time of different software tools on mt. DNA datasets

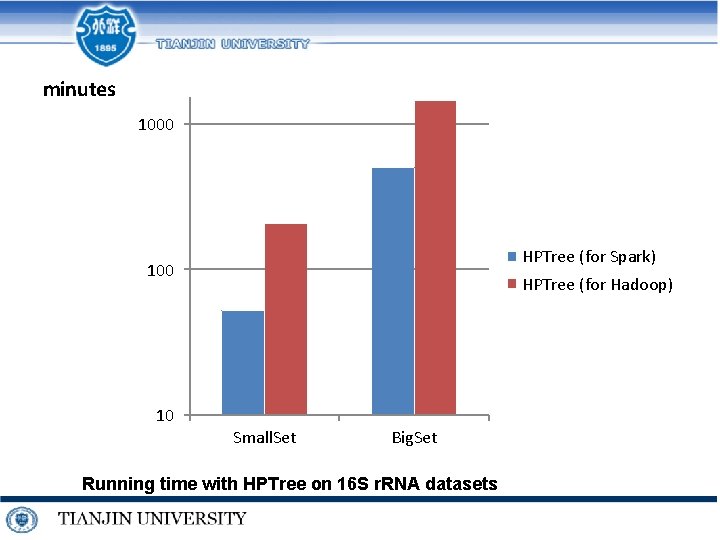
minutes 1000 HPTree (for Spark) 100 10 HPTree (for Hadoop) Small. Set Big. Set Running time with HPTree on 16 S r. RNA datasets


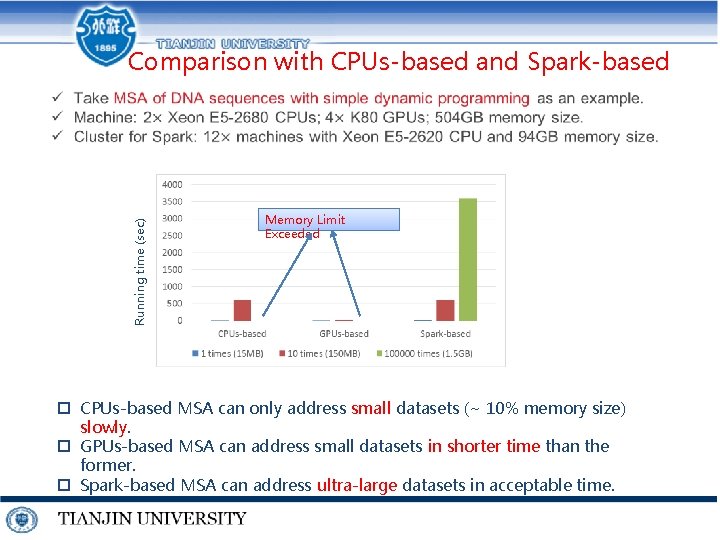
Running time (sec) Comparison with CPUs-based and Spark-based Memory Limit Exceeded p CPUs-based MSA can only address small datasets (~ 10% memory size) slowly. p GPUs-based MSA can address small datasets in shorter time than the former. p Spark-based MSA can address ultra-large datasets in acceptable time.

Software http: //lab. malab. cn/soft/halign/


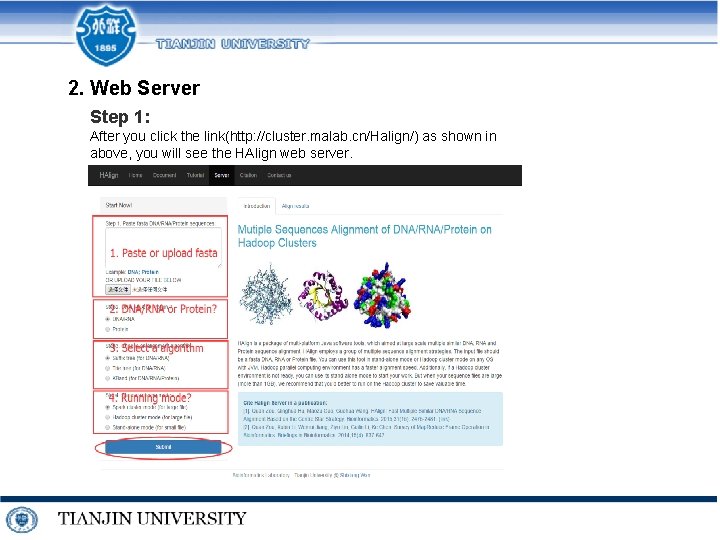
2. Web Server Step 1: After you click the link(http: //cluster. malab. cn/Halign/) as shown in above, you will see the HAlign web server.

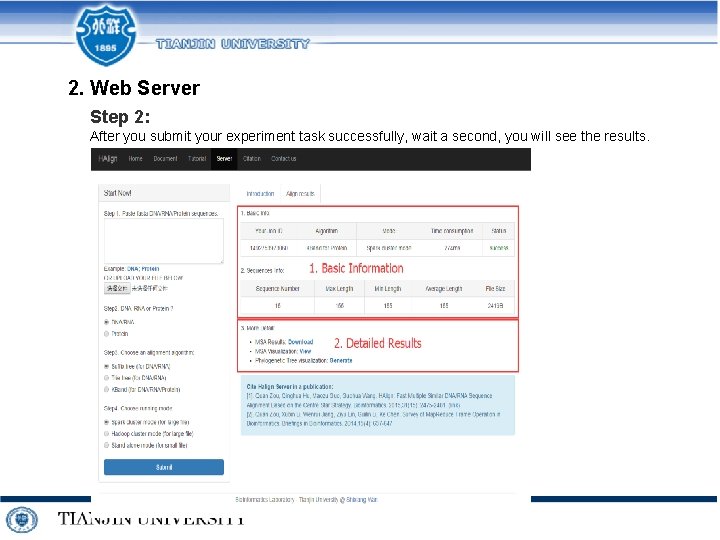
2. Web Server Step 2: After you submit your experiment task successfully, wait a second, you will see the results.

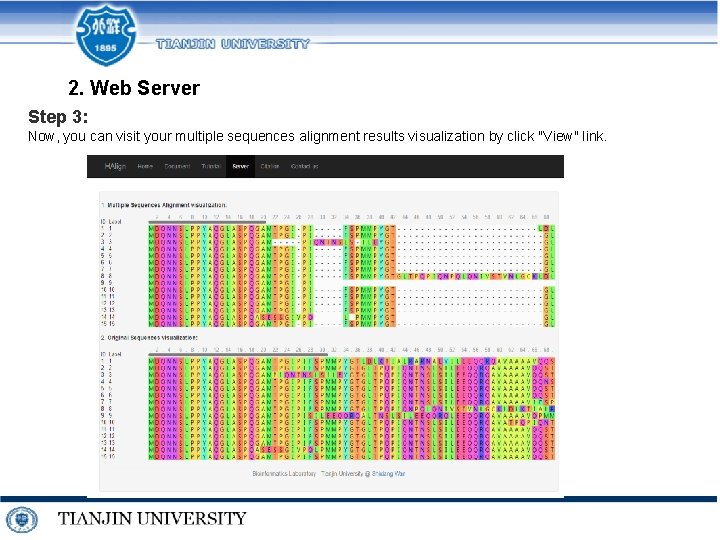
2. Web Server Step 3: Now, you can visit your multiple sequences alignment results visualization by click "View" link.

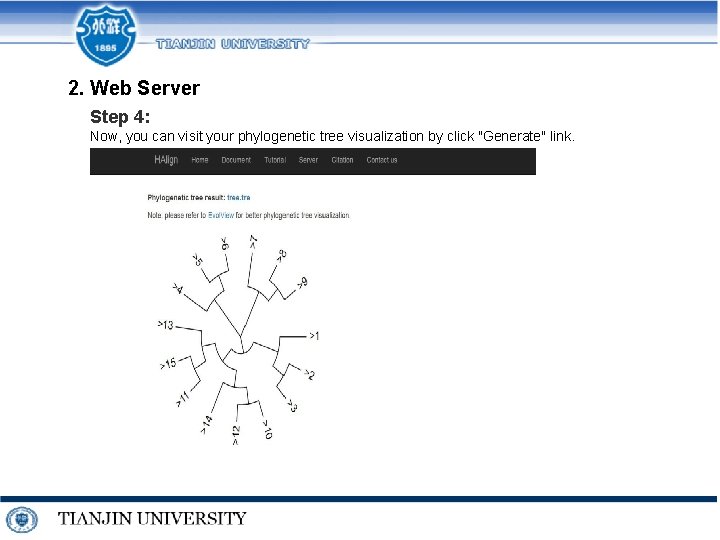
2. Web Server Step 4: Now, you can visit your phylogenetic tree visualization by click "Generate" link.

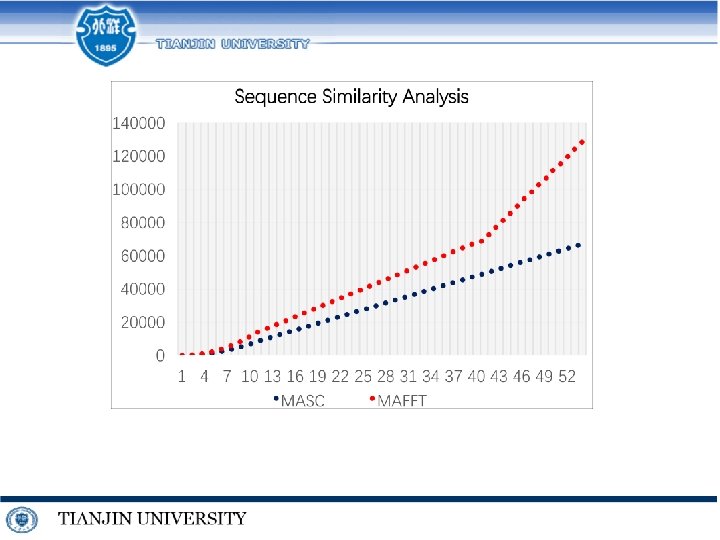
References on MSA • Quan Zou, Qinghua Hu, Maozu Guo, Guohua Wang. HAlign: Fast Multiple Similar DNA/RNA Sequence Alignment Based on the Centre Star Strategy. Bioinformatics. 2015, 31(15): 2475 -2481 • Xi Chen, Chen Wang, Shanjiang Tang, Ce Yu, Quan Zou. CMSA: A heterogeneous CPU/GPU computing system for multiple similar RNA/DNA sequence alignment. BMC Bioinformatics. 2017, 18: 315 • Shixiang Wan, Quan Zou*. HAlign-II: efficient ultra-large multiple sequence alignment and phylogenetic tree reconstruction with distributed and parallel computing. Algorithms for Molecular Biology. 2017, 12: 25 • Wenhe Su, Quan Zou, etc. MASC: A Linear Method for Multiple Nucleotide Sequence Alignment on Spark Parallel Framework. Journal of Computational Biology. Accepted

Outline • Sequence alignment – Algorithm – Parallel • Identification and mining – micro. RNA – machine learning related works • Function prediction – mi. RNA disease relationship – crops yield related genes

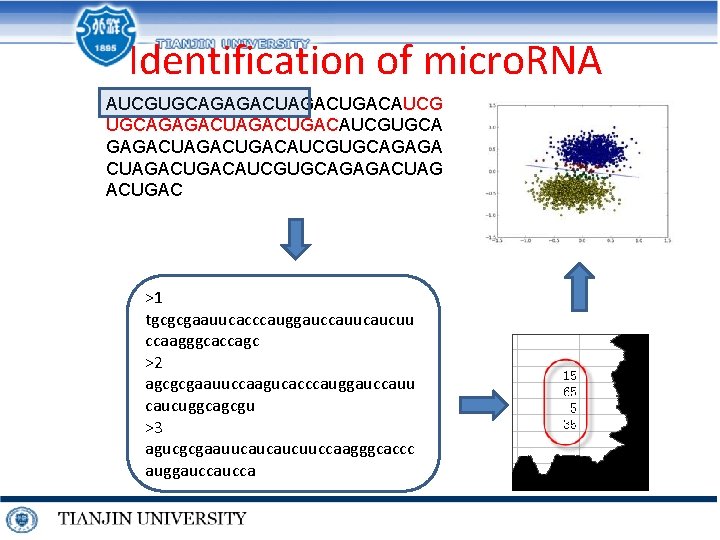
Identification of micro. RNA AUCGUGCAGAGACUGACAUCGUGCA GAGACUGACAUCGUGCAGAGA CUAGACUGACAUCGUGCAGAGACUAG ACUGAC >1 tgcgcgaauucacccauggauccauucaucuu ccaagggcaccagc >2 agcgcgaauuccaagucacccauggauccauu caucuggcagcgu >3 agucgcgaauucaucaucuuccaagggcaccc auggaucca

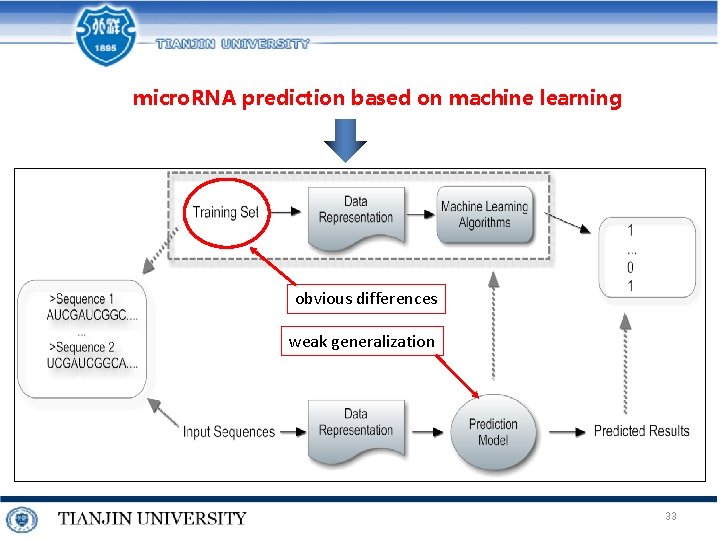
micro. RNA prediction based on machine learning obvious differences weak generalization 33

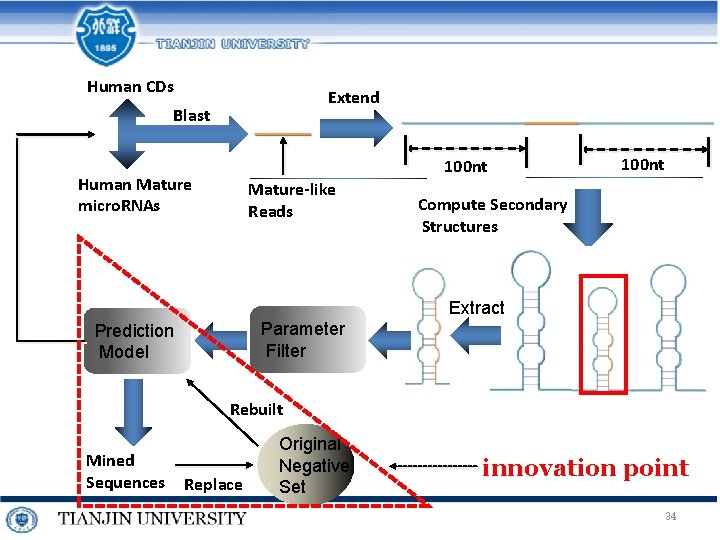
Human CDs Extend Blast 100 nt Human Mature micro. RNAs Mature-like Reads 100 nt Compute Secondary Structures Extract Parameter Filter Prediction Model Rebuilt Mined Sequences Replace Original Negative Set innovation point 34

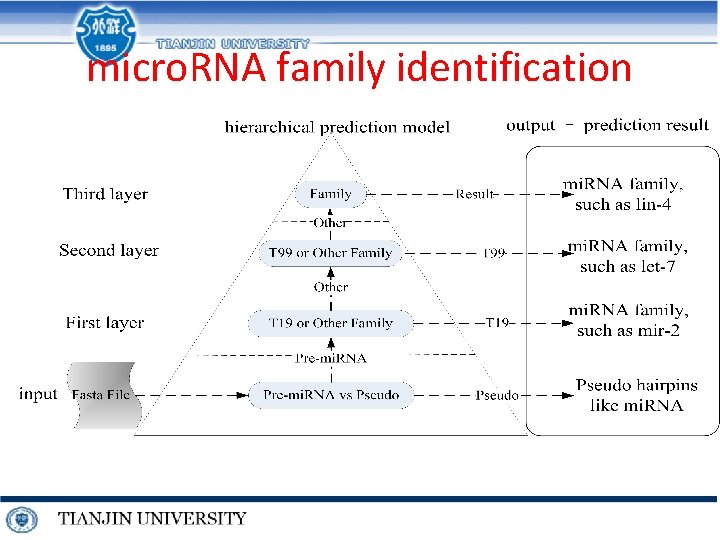
micro. RNA family identification

http: //lab. malab. cn/~wly/mirna. Detect. html 2021/10/20 36/30

Novel mi. RNA found by our method 1 37/30

Dinoflagellates genome (甲藻) Lin, et al. The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science. 2015, 350(6261): 691 -694.

Outline • Sequence alignment – Algorithm – Parallel • Identification and mining – micro. RNA – machine learning related works • Function prediction – mi. RNA disease relationship – crops yield related genes

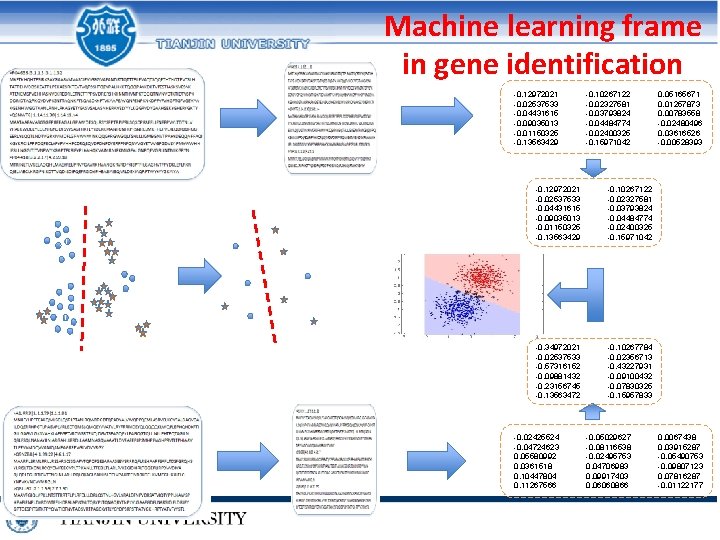
Machine learning frame in gene identification -0. 12972021 -0. 02537533 -0. 04431615 -0. 09035013 -0. 01150325 -0. 13563429 -0. 10267122 -0. 02327581 -0. 03793824 -0. 04484774 -0. 02400325 -0. 15971042 -0. 34972021 -0. 02537533 -0. 57316152 -0. 09881432 -0. 23156745 -0. 13563472 -0. 10267784 -0. 02356713 -0. 43227931 -0. 09100432 -0. 07830325 -0. 15957833 -0. 02425524 -0. 04724623 0. 05580992 0. 0361518 0. 10447804 0. 11267566 -0. 05029627 -0. 08116538 -0. 02495753 0. 04706983 0. 09917403 0. 06060866 0. 05165671 0. 01257873 0. 00783558 -0. 02480496 0. 03616526 -0. 00528393 0. 0067438 0. 03915287 -0. 05490753 -0. 09807123 0. 07816287 -0. 01122177

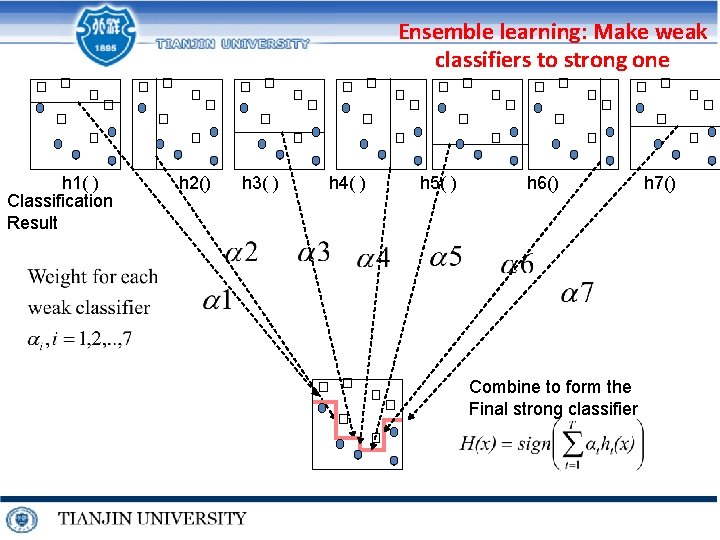
Ensemble learning: Make weak classifiers to strong one h 1( ) Classification Result h 2() h 3( ) h 4( ) h 5( ) h 6() Combine to form the Final strong classifier h 7()

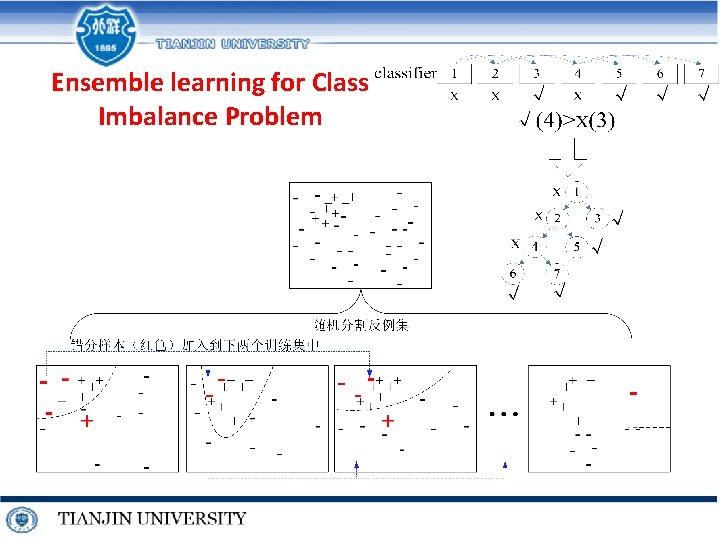
Ensemble learning for Class Imbalance Problem

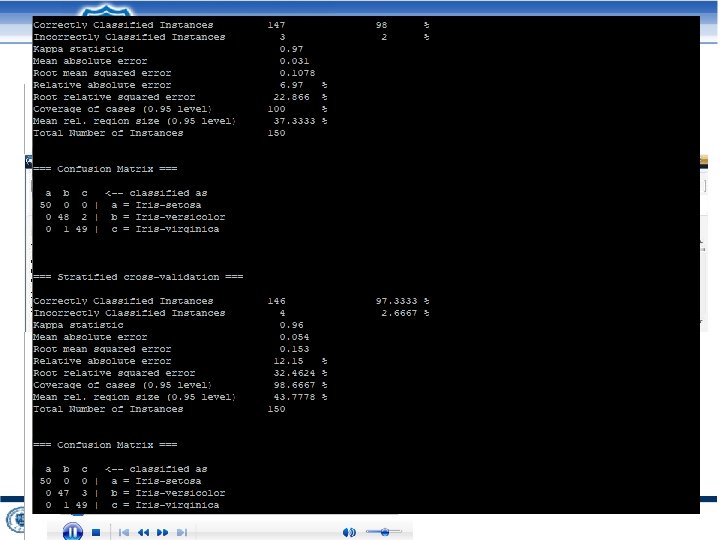
http: //lab. malab. cn/soft/Lib. D 3 C/ 2021/10/20

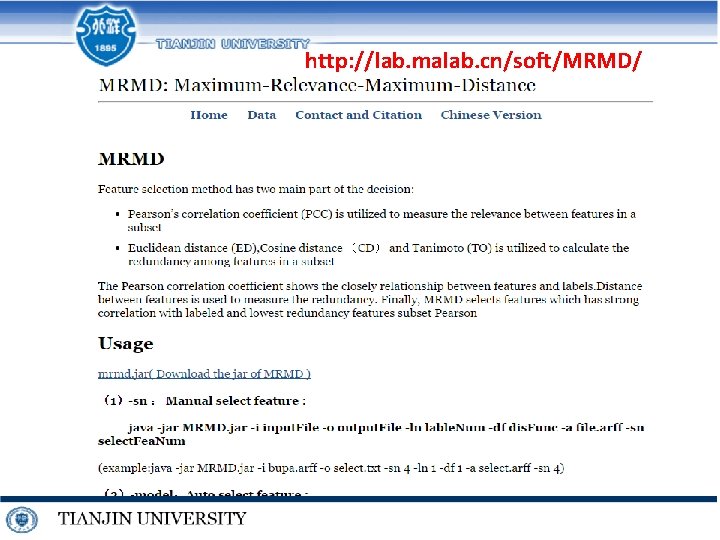
http: //lab. malab. cn/soft/MRMD/

Application in Bioinformatics • DNA Binding proteins – Li Song, Dapeng Li, Xiangxiang Zeng, Yunfeng Wu, Li Guo*, Quan Zou*. n. DNA-prot: Identification of DNA-binding Proteins Based on Unbalanced Classification. BMC Bioinformatics. 2014, 15: 298. • t. RNA – Quan Zou, et al. Improving t. RNAscan-SE annotation results via ensemble classifiers. Molecular Informatics. 2015, 34(11 -12): 761 -770 • mi. RNA – Leyi Wei, Minghong Liao, Yue Gao, Rongrong Ji, Zengyou He*, Quan Zou*. Improved and Promising Identification of Human Micro. RNAs by Incorporating a High-quality Negative Set. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2014, 11(1): 192 -201 • circle. RNA – Xiangxiang Zeng, Wei Lin, Maozu Guo, Quan Zou*. A comprehensive overview and evaluation of circular RNA detection tools. PLo. S Computational Biology. 2017, 13(6): e 1005420


zouquan@tju. edu. cn


References • Leyi Wei, Minghong Liao, Yue Gao, Rongrong Ji, Zengyou He*, Quan Zou*. Improved and Promising Identification of Human Micro. RNAs by Incorporating a High-quality Negative Set. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2014, 11(1): 192 -201 • Quan Zou*, Yaozong Mao, Lingling Hu, Yunfeng Wu, Zhiliang Ji*. mi. RClassify: An advanced web server for mi. RNA family classification and annotation. Computers in Biology and Medicine. 2014, 45: 157 -160 • Chen Lin, Wenqiang Chen, Cheng Qiu, Yunfeng Wu, Sridhar Krishnan, Quan Zou*. Lib. D 3 C: Ensemble Classifiers with a Clustering and Dynamic Selection Strategy. Neurocomputing. 2014, 123: 424 -435. • Quan Zou, Jiancang Zeng, Liujuan Cao, Rongrong Ji. A Novel Features Ranking Metric with Application to Scalable Visual and Bioinformatics Data Classification. Neurocomputing. 2016, 173: 346 -354

Outline • Sequence alignment – Algorithm – Parallel • Identification and mining – micro. RNA – machine learning related works • Function prediction – mi. RNA disease relationship – crops yield related genes


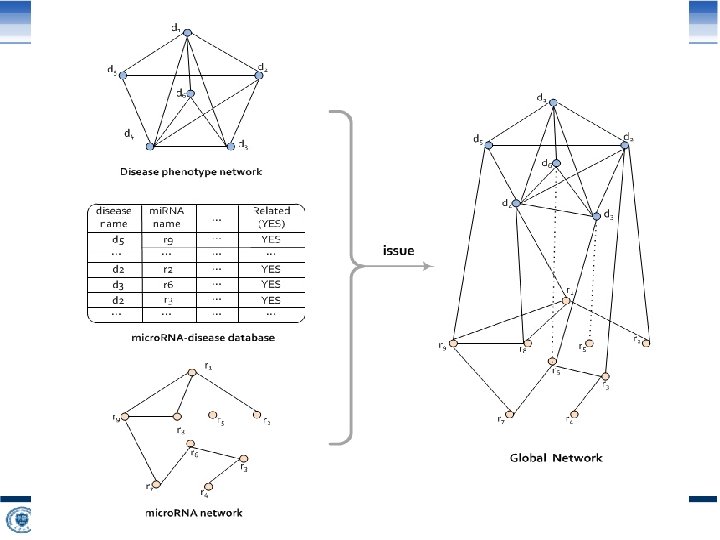
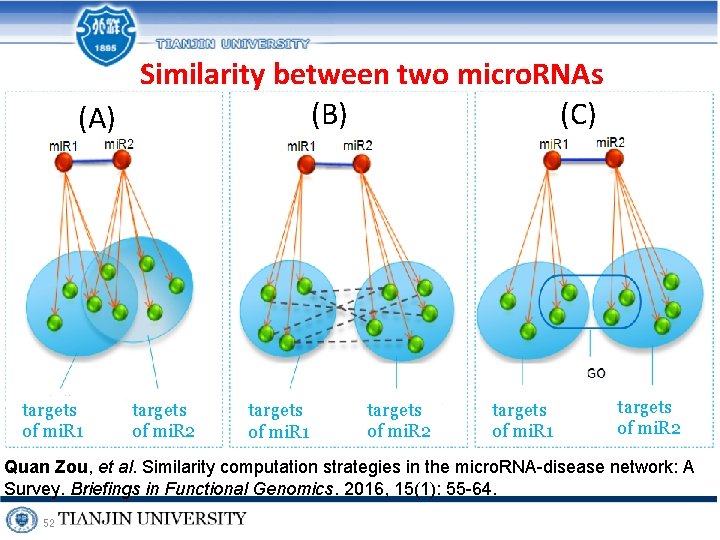
Similarity between two micro. RNAs (B) (C) (A) targets of mi. R 1 targets of mi. R 2 Quan Zou, et al. Similarity computation strategies in the micro. RNA-disease network: A Survey. Briefings in Functional Genomics. 2016, 15(1): 55 -64. 52

Wei Tang, Zhijun Liao, Quan Zou*. Which statistical significance test best detects oncomi. RNAs in cancer tissues? An exploratory analysis. Oncotarget. DOI: 10. 18632/oncotarget. 12828. 53

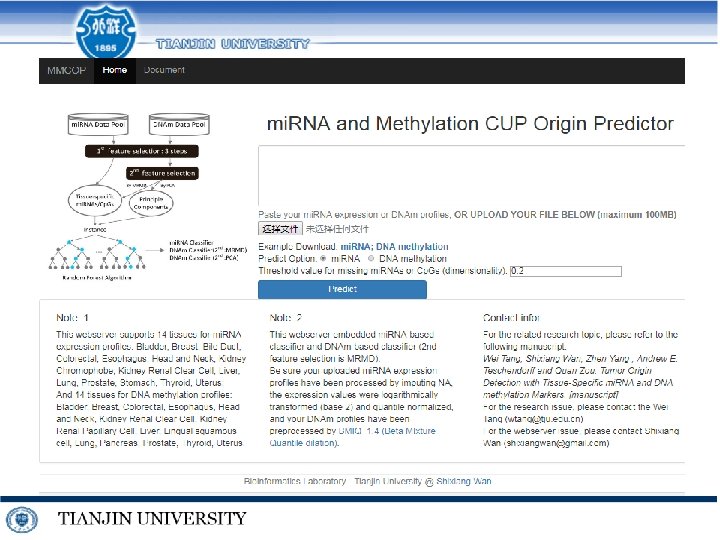
http: //lab. malab. cn/soft/MDPredict/

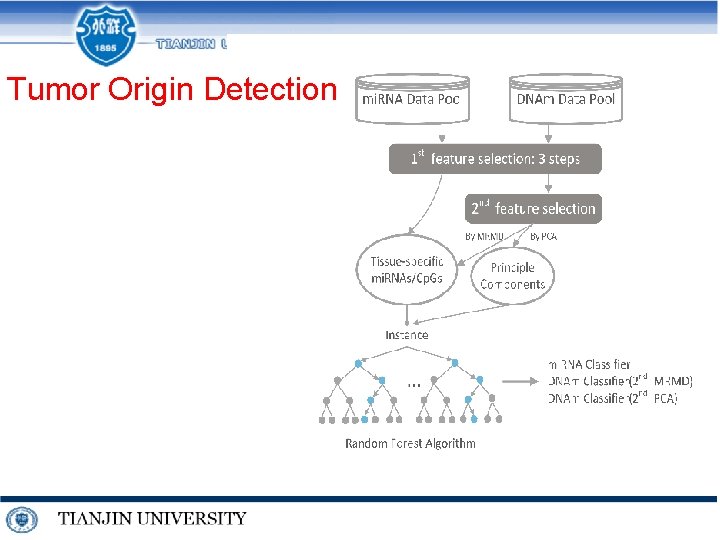
Tumor Origin Detection


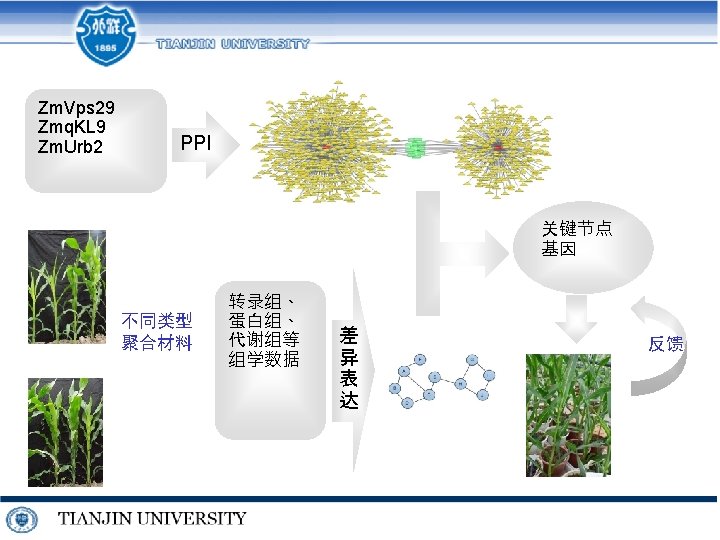
Outline • Sequence alignment – Algorithm – Parallel • Identification and mining – micro. RNA – machine learning related works • Function prediction – mi. RNA disease relationship – crops yield related genes

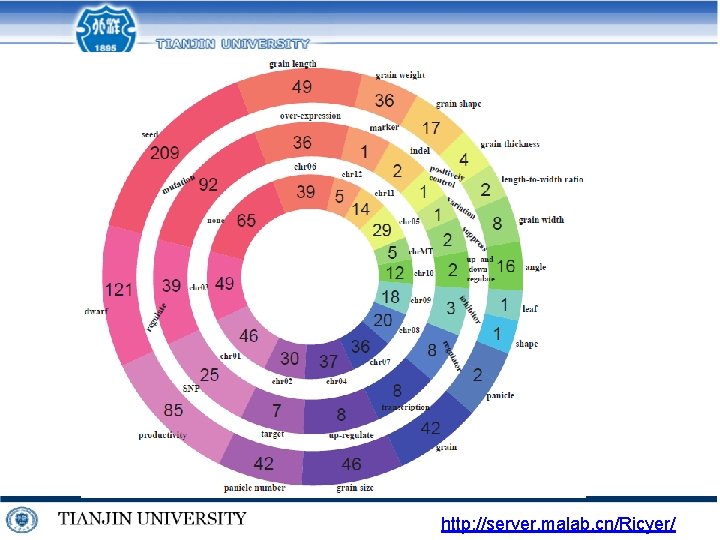
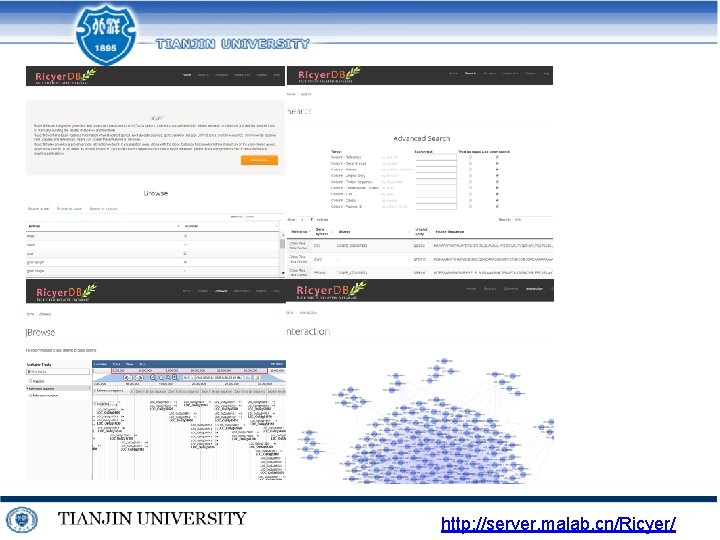
http: //server. malab. cn/Ricyer/

http: //server. malab. cn/Ricyer/

http: //server. malab. cn/Ricyer/


References • Xiangxiang Zeng, Xuan Zhang, Quan Zou*. Integrative approaches for predicting micro. RNA function and prioritizing disease-related micro. RNA using biological interaction networks. Briefings in Bioinformatics. 2016, 17(2): 193 -203. • Yuansheng Liu, Xiangxiang Zeng, Zengyou He*, Quan Zou*. Inferring micro. RNA-disease associations by random walk on a heterogeneous network with multiple data sources. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2017, 14(4): 905 -915 • Wei Tang, Zhijun Liao, Quan Zou*. Which statistical significance test best detects oncomi. RNAs in cancer tissues? An exploratory analysis. Oncotarget. 2016, 7(51): 85613 -85623 • Wei Tang, Shixiang Wan, Zhen Yang, Andrew E. Teschendorff*, Quan Zou*. Tumor Origin Detection with Tissue-Specific mi. RNA and DNA methylation Markers. Bioinformatics. Doi: 10. 1093/bioinformatics/btx 622
