Osteochondromagenesis Loss of heterozygosity modeled via Cremediated inversion




















- Slides: 34

Osteochondromagenesis: Loss of heterozygosity modeled via Cre-mediated inversion of the second exon of Ext 1 in chondrocytes Department of Orthopaedics and Rehabilitation University of Iowa Hospitals & Clinics Kevin B. Jones, MD Charles Searby, BS Gail Kurriger, BS James Martin, Ph. D Peter J. Roughley, Ph. D Jose A. Morcuende, MD, Ph. D Joseph A. Buckwalter, MD, MS Val C. Sheffield, MD, Ph. D Connective Tissue Oncology Society London, UK Friday, 14 November 2008

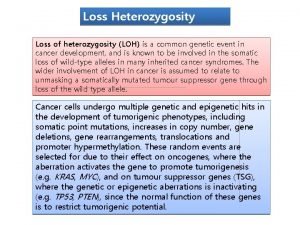
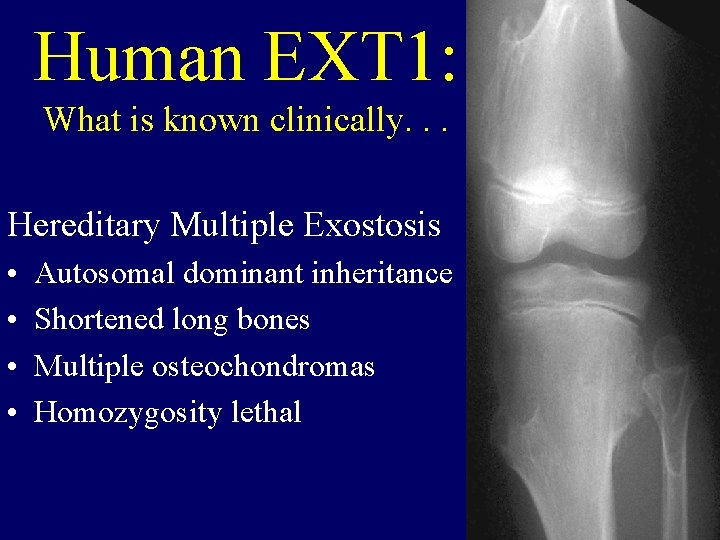
Human EXT 1: What is known clinically. . . Hereditary Multiple Exostosis • • Autosomal dominant inheritance Shortened long bones Multiple osteochondromas Homozygosity lethal


EXT 1 & EXT 2: Tumor Supressor Genes? • Loss of heterozygosity in chondrocytes of cartilage cap of HME osteochondromas • Both alleles somatically mutated in some solitary osteochondromas Raskind et al. 1995; Bovee et al. 1999; Hecht et al. 1995 and 1997.


Ext 1 Knock-Out Mice by Lin et al. 2000 Mouse Ext 1 has 99% Homology to Human EXT 1 PROBLEMS: • Homozygous Ext 1 -knock-out mice do not survive to birth • Heterozygous Ext 1 -knock-out mice very rarely form osteochondromas Simlar with Ext 2 knock-out mice, Sitckens et al. 2005.

Of Mice and Men

Of Mice and Men

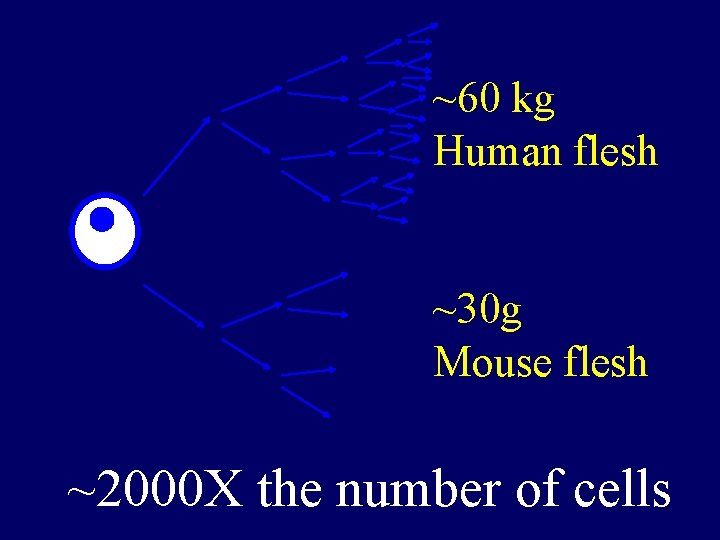
~60 kg Human flesh ~30 g Mouse flesh ~2000 X the number of cells

Haploinsufficiency or Loss of Heterozygosity?

Methods

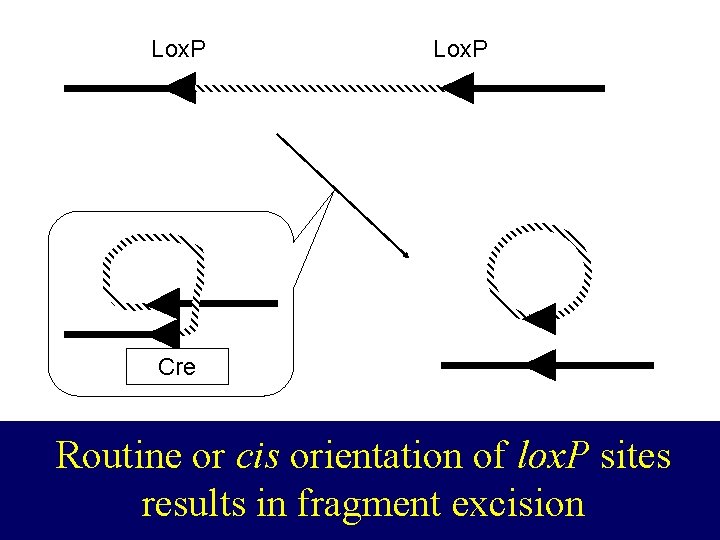
lox. P Lox. P CRE Cre Routine or cis orientation of lox. P sites results in fragment excision

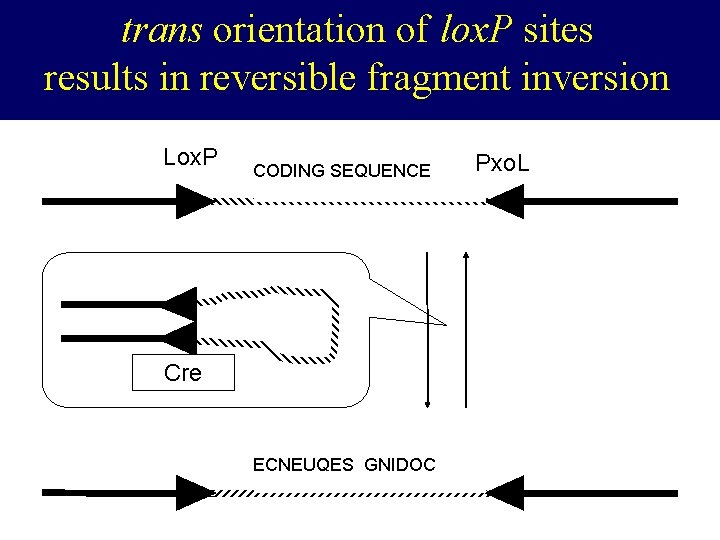
trans orientation of lox. P sites results in reversible fragment inversion Lox. P Pxol CODING SEQUENCE CRE Cre ECNEUQES GNIDOC Pxo. L lox. P

You do the math. . . • Cre-mediated inversion yields 40% reverse orientation per trans-floxed allele • Homozygosity for trans-floxed alleles should yield 40% loss of total copies of functional gene across tissue – – 20% of cells will remain unaffected and fwd/fwd 20% of cells will end up fwd/fwd after inversion 40% of cells will end up fwd/rev after inversion 20% of cells will end up rev/rev after inversion

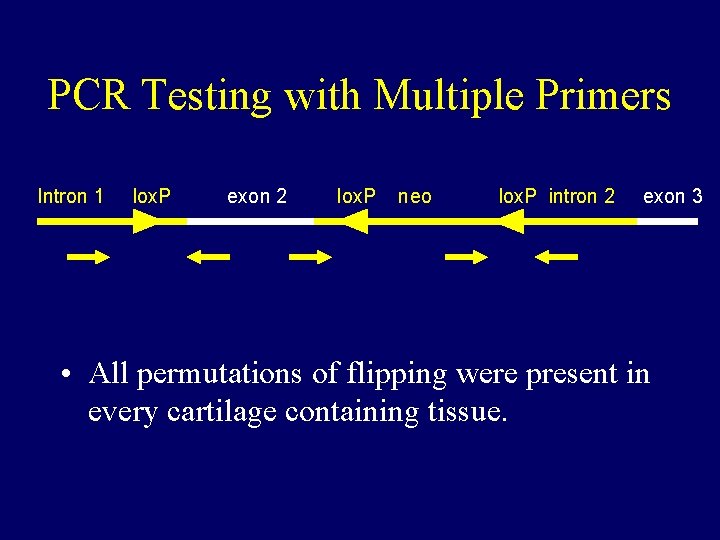
Targeting Construct Intron 1 lox. P exon 2 lox. P neo lox. P intron 2 • Exon 2 contains two of the very few diseasecausing missense mutations at highly conserved amino acid residues 339 and 340. • Exon 2 inversion not only disrupts the sequence, but introduces a stop codon in the reading frame exon 3

Cre-Recombinase Driver • Doxycycline-inducible Collagen type II promoter-Cre • Doxycycline adminstered via maternal drinking water during second week of life Gover and Roughley et al. , 2006

Results

PCR Testing with Multiple Primers Intron 1 lox. P exon 2 lox. P neo lox. P intron 2 exon 3 • All permutations of flipping were present in every cartilage containing tissue.

excised neo exon 2 excised neo reversed neo 2 exon excised neo reversed exon 2 neo excised neo reversed exon 2

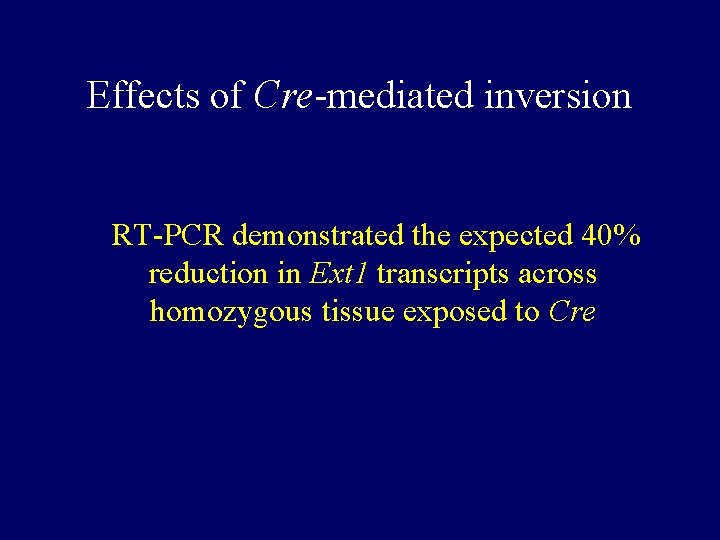
Effects of Cre-mediated inversion RT-PCR demonstrated the expected 40% reduction in Ext 1 transcripts across homozygous tissue exposed to Cre

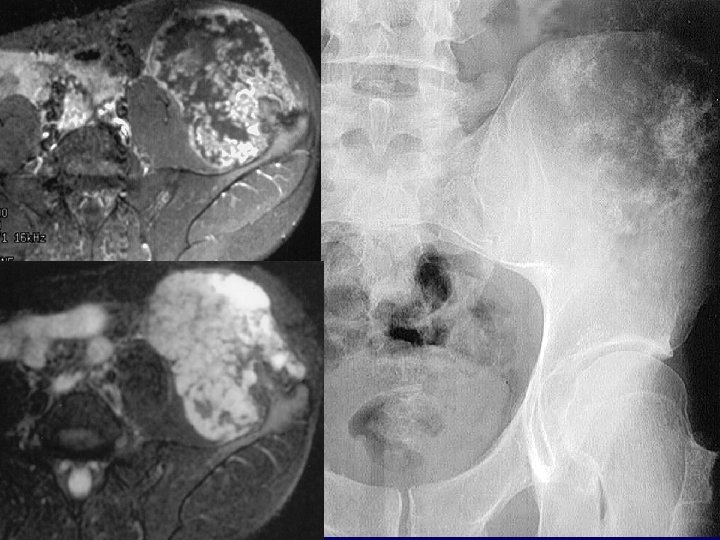
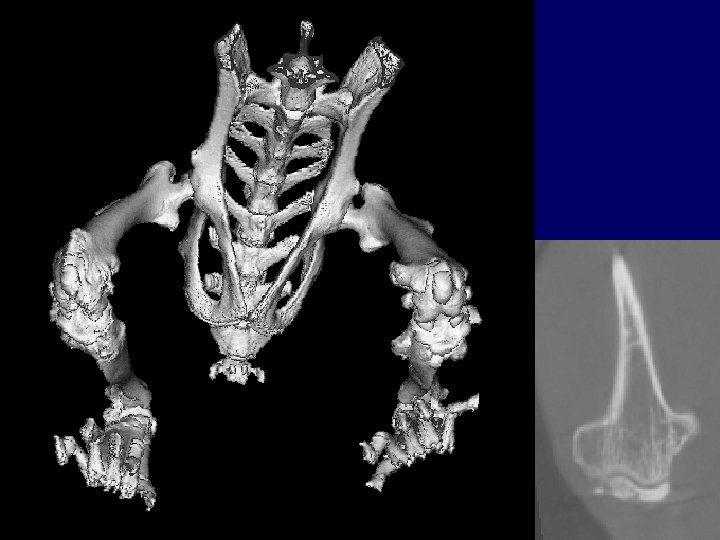
Phenotype: Osteochondromas?

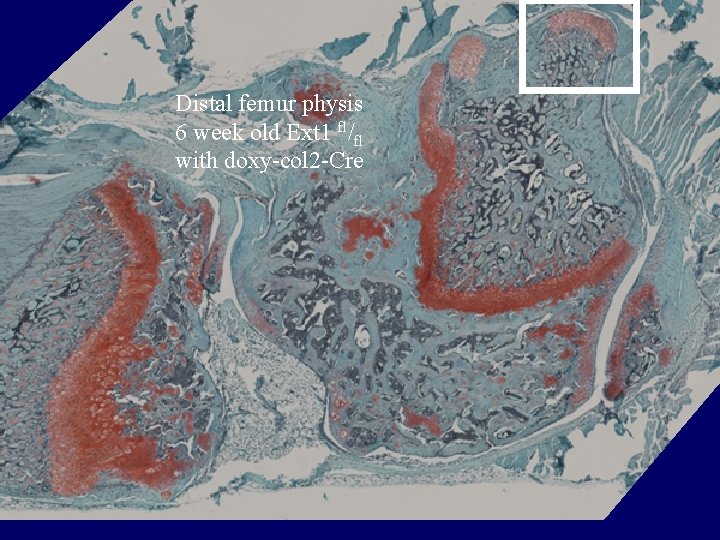
Distal femur physis 6 week old Ext 1 fl/fl with doxy-col 2 -Cre


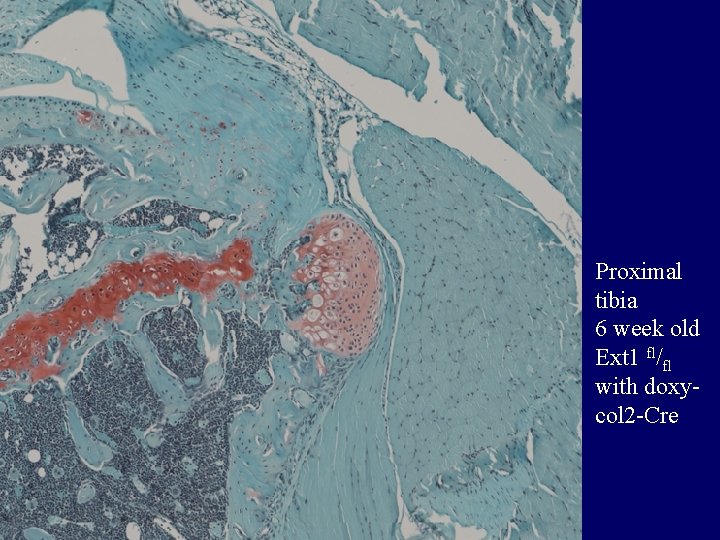
Proximal tibia 6 week old Ext 1 fl/fl with doxycol 2 -Cre


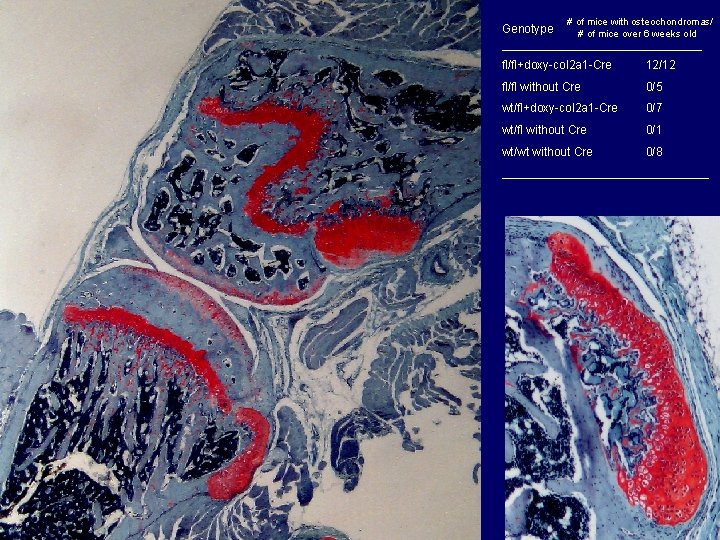
Genotype # of mice with osteochondromas/ # of mice over 6 weeks old _______________________ fl/fl+doxy-col 2 a 1 -Cre 12/12 fl/fl without Cre 0/5 wt/fl+doxy-col 2 a 1 -Cre 0/7 wt/fl without Cre 0/1 wt/wt without Cre 0/8 ________________

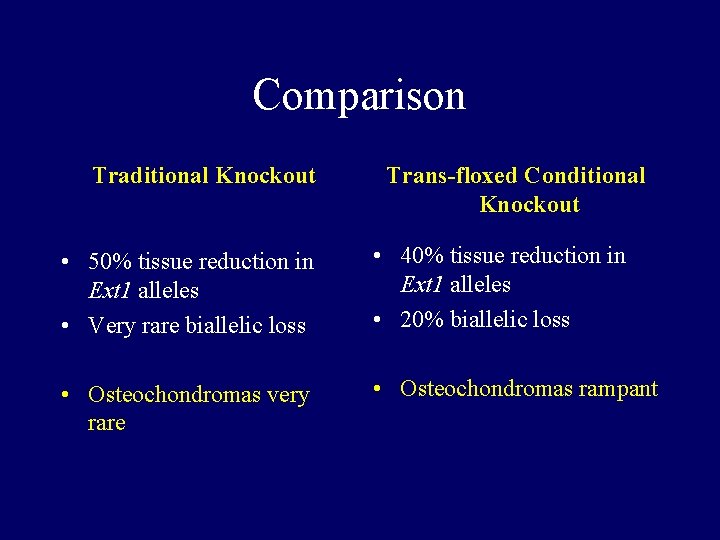
Comparison Traditional Knockout Trans-floxed Conditional Knockout • 50% tissue reduction in Ext 1 alleles • Very rare biallelic loss • 40% tissue reduction in Ext 1 alleles • 20% biallelic loss • Osteochondromas very rare • Osteochondromas rampant

Discussion Loss of heterozygosity is the mechanism of osteochondromagenesis in the setting of hereditary multiple exostoses

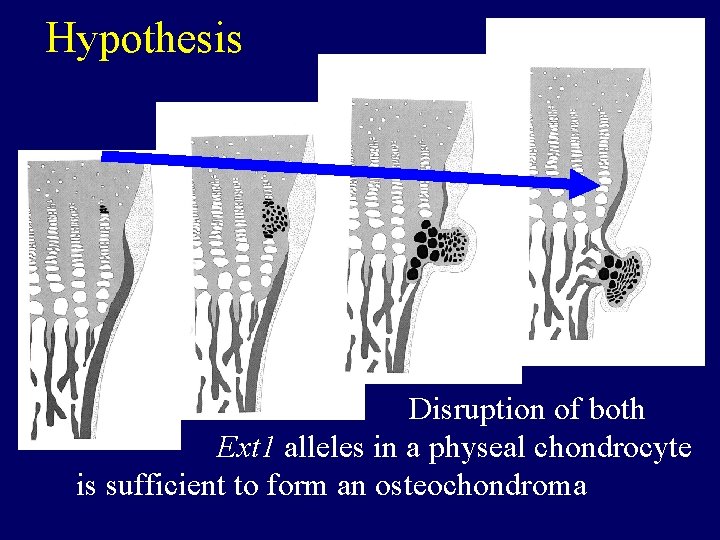
Hypothesis Disruption of both Ext 1 alleles in a physeal chondrocyte is sufficient to form an osteochondroma

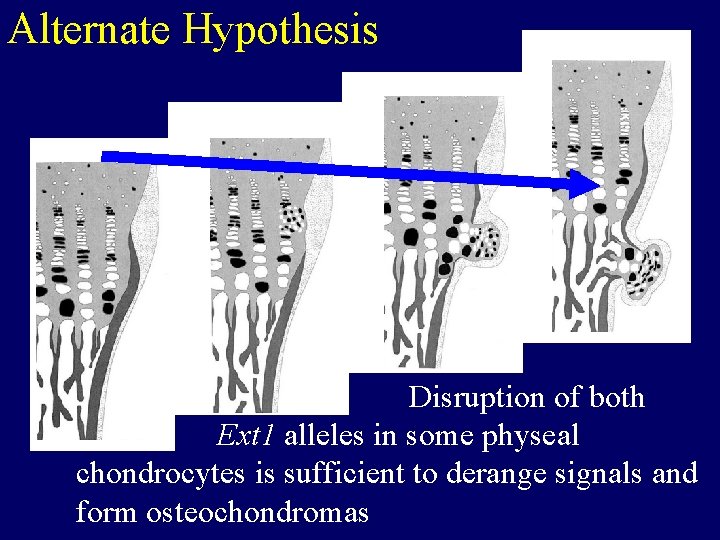
Alternate Hypothesis Disruption of both Ext 1 alleles in some physeal chondrocytes is sufficient to derange signals and form osteochondromas

Hypothesis vs. Alternate Hypothesis Are osteochondromas clonal (at least consistent) for reverse orientation of exon 2? On-going work with laser capture micro-dissection and in situ hybridization

Conclusion Osteochondromageneis resulted from lowprevalence biallelic disruption of Ext 1 in chondrocytes, modeling loss of heterozygosity with a unique twist on the standard conditional knock-out

Project Funding Support • OREF Resident Research Award 2003 • Department of Orthopaedics and Rehabilitation, University of Iowa • Val C. Sheffield Genetics Laboratory Department of Orthopaedics and Rehabilitation University of Iowa Hospitals & Clinics

Osteochondromagenesis: Loss of heterozygosity modeled via Cre-mediated inversion of the second exon of Ext 1 in chondrocytes Department of Orthopaedics and Rehabilitation University of Iowa Hospitals & Clinics Kevin B. Jones, MD Charles Searby, BS Gail Kurriger, BS James Martin, Ph. D Peter J. Roughley, Ph. D Jose A. Morcuende, MD, Ph. D Joseph A. Buckwalter, MD, MS Val C. Sheffield, MD, Ph. D Connective Tissue Oncology Society London, UK Friday, 14 November 2008
 Loss of heterozygosity
Loss of heterozygosity Loss of heterozygosity slideshare
Loss of heterozygosity slideshare Unit cost meaning
Unit cost meaning Que significa el via lucis
Que significa el via lucis Sistema piramidal y extrapiramidal
Sistema piramidal y extrapiramidal Decimoquinta estacion via crucis
Decimoquinta estacion via crucis Via erudita e via popular
Via erudita e via popular La via negativa
La via negativa The biography of langston hughes
The biography of langston hughes Elizabethan tragedies were modeled on plays from
Elizabethan tragedies were modeled on plays from The area of a rectangular fountain is (x^2+12x+20)
The area of a rectangular fountain is (x^2+12x+20) Example of point in geometry
Example of point in geometry The path of a placekicked football can be modeled
The path of a placekicked football can be modeled Which sum or difference is modeled by the algebra tiles
Which sum or difference is modeled by the algebra tiles Arbol de problemas de un proyecto de inversion
Arbol de problemas de un proyecto de inversion Covering of spermatic cord
Covering of spermatic cord Inversion and fronting
Inversion and fronting Arbol de problemas de un proyecto de inversion
Arbol de problemas de un proyecto de inversion Una inversión consiste en
Una inversión consiste en Vg
Vg Stimulated emission
Stimulated emission Nipple retraction or inversion
Nipple retraction or inversion Inversion in thanatopsis
Inversion in thanatopsis Inversion publica chile
Inversion publica chile Inversion
Inversion Inversion hotline
Inversion hotline 47, xx, +21
47, xx, +21 Inversión forzosa de no menos del 1
Inversión forzosa de no menos del 1 Arbol de problemas de un proyecto de inversion
Arbol de problemas de un proyecto de inversion How to calculate inverse of a matrix
How to calculate inverse of a matrix Paracentric vs pericentric inversion
Paracentric vs pericentric inversion Inversion dna
Inversion dna Inversion after negative adverbs
Inversion after negative adverbs Inversion mutation
Inversion mutation Inversion of control
Inversion of control