Orthology Paralogy etc Orthologs Two genes each from







- Slides: 25

Orthology & Paralogy (etc. ) Orthologs: Two genes, each from a different species, that descended from a single common ancestral gene (note no regard to function! and does NOT require one-to-one relationships) Paralogs: Two or more genes, often thought of as within the same species, that originated by one or more gene duplication events 1

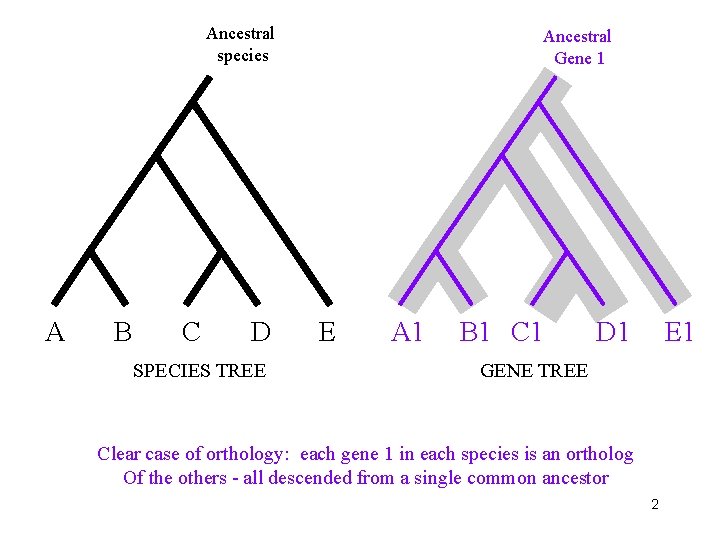
Ancestral species A B C D SPECIES TREE Ancestral Gene 1 E A 1 B 1 C 1 D 1 E 1 GENE TREE Clear case of orthology: each gene 1 in each species is an ortholog Of the others - all descended from a single common ancestor 2

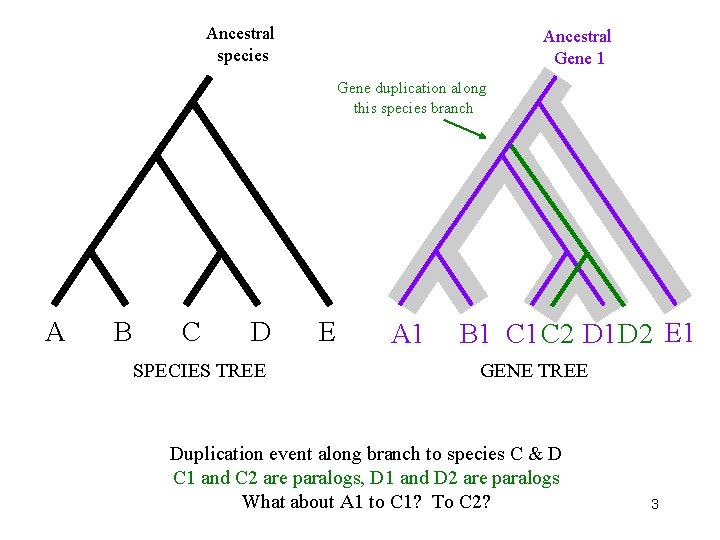
Ancestral species Ancestral Gene 1 Gene duplication along this species branch A B C D SPECIES TREE E A 1 B 1 C 2 D 1 D 2 E 1 GENE TREE Duplication event along branch to species C & D C 1 and C 2 are paralogs, D 1 and D 2 are paralogs What about A 1 to C 1? To C 2? 3

Orthology & Paralogy (etc. ) Orthologs: Two genes, each from a different species, that descended from a single common ancestral gene (note no regard to function!) Paralogs: Two or more genes, within the same species, that originated by one or more gene duplication events Also now many subtle variants: Outparalogs: cross-species paralogs (i. e. gene duplication BEFORE speciation) Inparalogs: lineage-specific duplication (i. e. duplication AFTER speciation) Ohnolog: duplicates originating from a whole-genome duplication (WGD) Xenolog: genes related by horizontal gene transfer between species 4

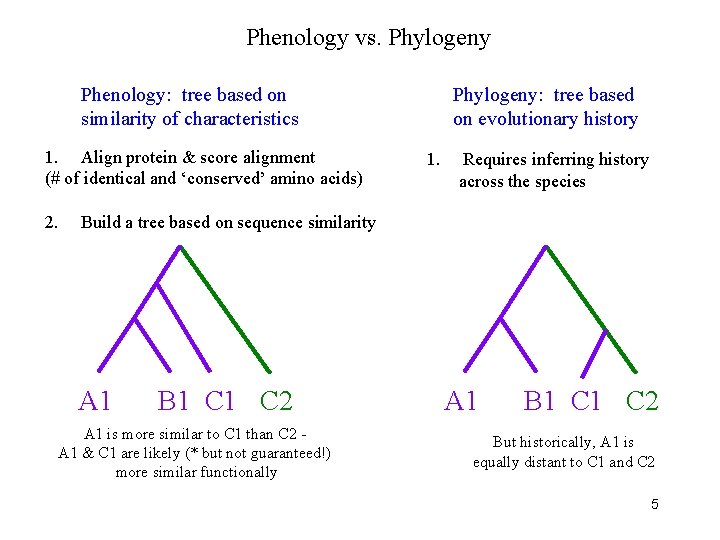
Phenology vs. Phylogeny Phenology: tree based on similarity of characteristics 1. Align protein & score alignment (# of identical and ‘conserved’ amino acids) 2. Phylogeny: tree based on evolutionary history 1. Requires inferring history across the species Build a tree based on sequence similarity A 1 B 1 C 2 A 1 is more similar to C 1 than C 2 A 1 & C 1 are likely (* but not guaranteed!) more similar functionally A 1 B 1 C 2 But historically, A 1 is equally distant to C 1 and C 2 5

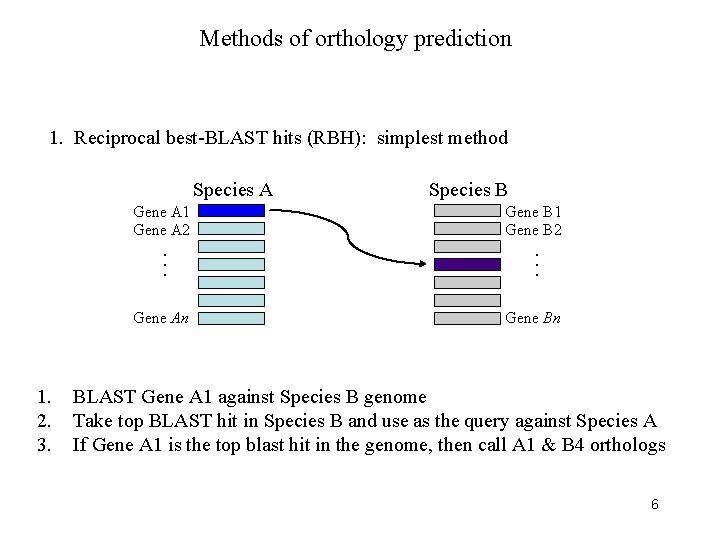
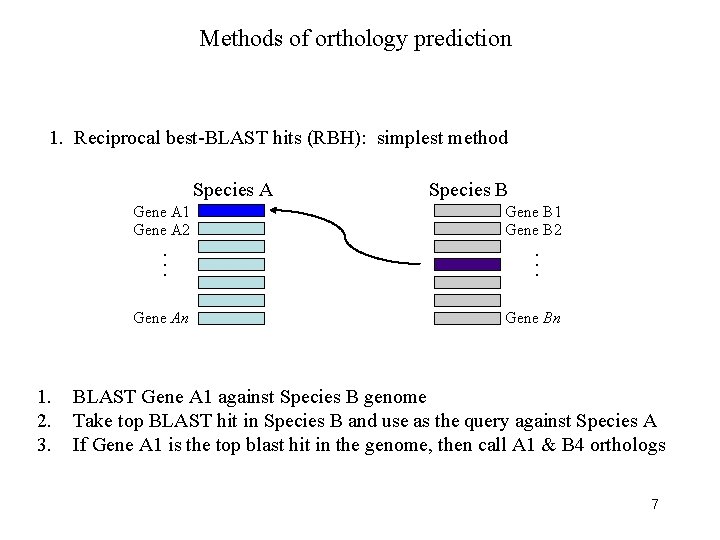
Methods of orthology prediction 1. Reciprocal best-BLAST hits (RBH): simplest method Species A Gene A 1 Gene A 2 1. 2. 3. Gene B 1 Gene B 2 . . . Gene An Species B Gene Bn BLAST Gene A 1 against Species B genome Take top BLAST hit in Species B and use as the query against Species A If Gene A 1 is the top blast hit in the genome, then call A 1 & B 4 orthologs 6

Methods of orthology prediction 1. Reciprocal best-BLAST hits (RBH): simplest method Species A Gene A 1 Gene A 2 1. 2. 3. Gene B 1 Gene B 2 . . . Gene An Species B Gene Bn BLAST Gene A 1 against Species B genome Take top BLAST hit in Species B and use as the query against Species A If Gene A 1 is the top blast hit in the genome, then call A 1 & B 4 orthologs 7

Problems with RBH * Clear cases where the top BLAST hit is NOT the ortholog e. g. top hits can be highly conserved common domains * Gene duplications in one species can completely obscure orthologous hits * Orthologs with very low sequence homology can be missed altogether 8

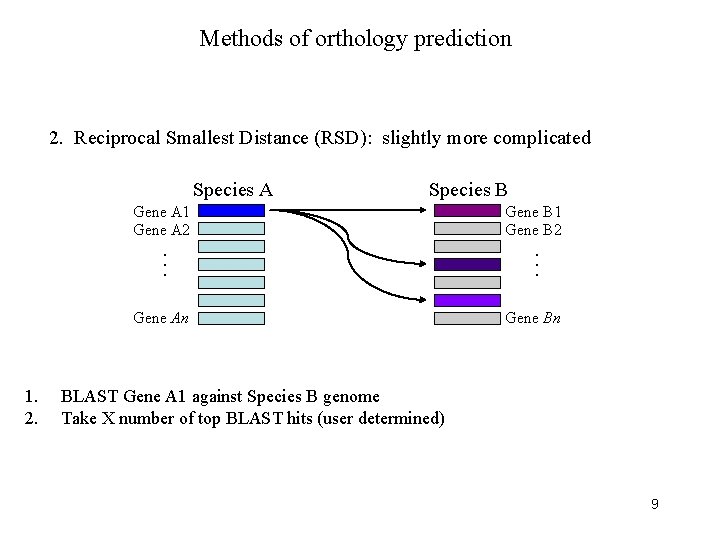
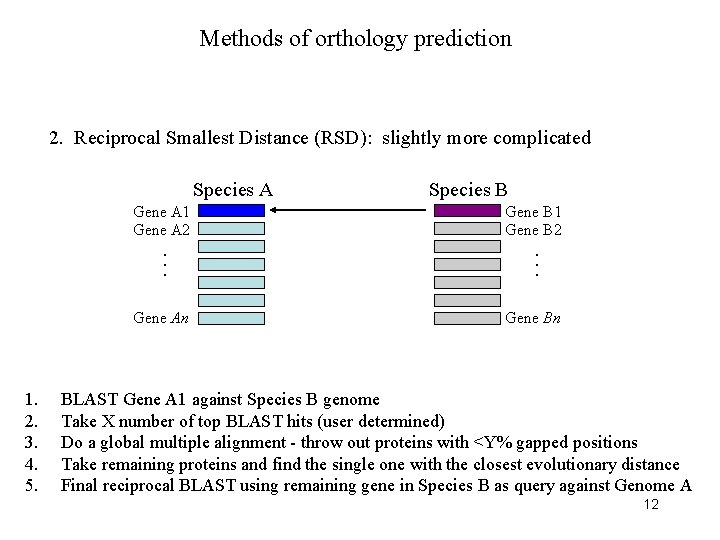
Methods of orthology prediction 2. Reciprocal Smallest Distance (RSD): slightly more complicated Species A Species B Gene A 1 Gene A 2 1. 2. . . . Gene An Gene B 1 Gene B 2 Gene Bn BLAST Gene A 1 against Species B genome Take X number of top BLAST hits (user determined) 9

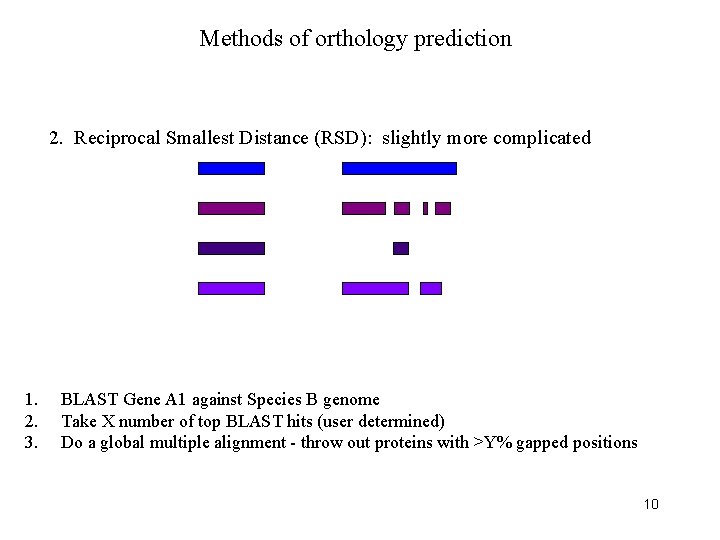
Methods of orthology prediction 2. Reciprocal Smallest Distance (RSD): slightly more complicated 1. 2. 3. BLAST Gene A 1 against Species B genome Take X number of top BLAST hits (user determined) Do a global multiple alignment - throw out proteins with >Y% gapped positions 10

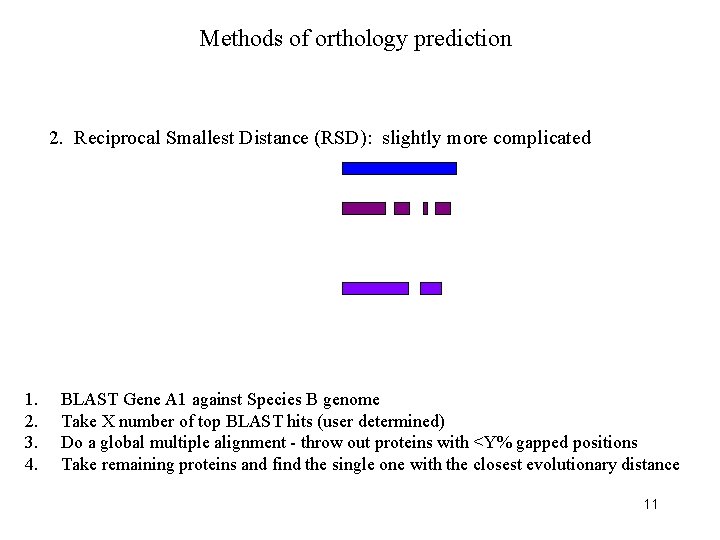
Methods of orthology prediction 2. Reciprocal Smallest Distance (RSD): slightly more complicated 1. 2. 3. 4. BLAST Gene A 1 against Species B genome Take X number of top BLAST hits (user determined) Do a global multiple alignment - throw out proteins with <Y% gapped positions Take remaining proteins and find the single one with the closest evolutionary distance 11

Methods of orthology prediction 2. Reciprocal Smallest Distance (RSD): slightly more complicated Species A Gene A 1 Gene A 2 1. 2. 3. 4. 5. Gene B 1 Gene B 2 . . . Gene An Species B Gene Bn BLAST Gene A 1 against Species B genome Take X number of top BLAST hits (user determined) Do a global multiple alignment - throw out proteins with <Y% gapped positions Take remaining proteins and find the single one with the closest evolutionary distance Final reciprocal BLAST using remaining gene in Species B as query against Genome A 12

Problems with RSD * Clear cases where the top BLAST hit is NOT the ortholog e. g. top hits can be highly conserved common domains * Gene duplications in one species can completely obscure orthologous hits * Orthologs with very low sequence homology can be missed altogether 13

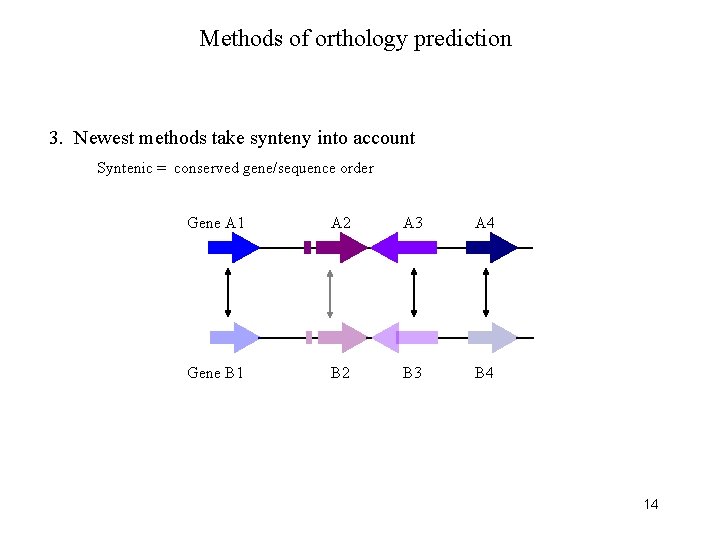
Methods of orthology prediction 3. Newest methods take synteny into account Syntenic = conserved gene/sequence order Gene A 1 A 2 A 3 A 4 Gene B 1 B 2 B 3 B 4 14

Problems with Synteny-based Methods * Clear cases where the top BLAST hit is NOT the ortholog e. g. top hits can be highly conserved common domains * Gene duplications in one species less likely to obscure things * Orthologs with low sequence homology not part of a larger duplication could still be missed 15

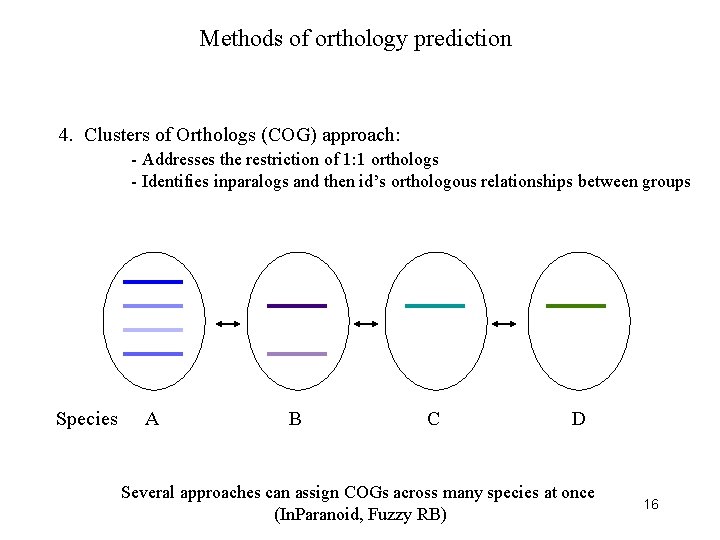
Methods of orthology prediction 4. Clusters of Orthologs (COG) approach: - Addresses the restriction of 1: 1 orthologs - Identifies inparalogs and then id’s orthologous relationships between groups Species A B C D Several approaches can assign COGs across many species at once (In. Paranoid, Fuzzy RB) 16

Lots of different databases of orthologs (esp. for model organisms)

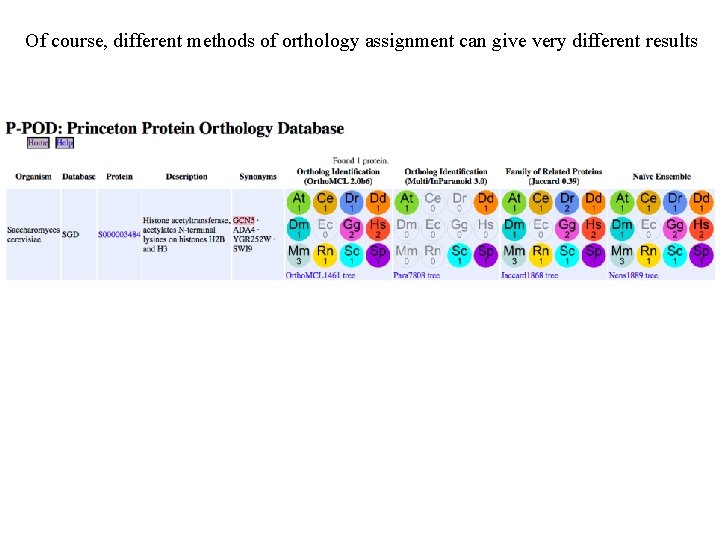
Of course, different methods of orthology assignment can give very different results

AND … genome errors can really obscure things Bad genome annotations can affect orthology & paralogy relationships - missing genes, fused genes, incorrect start/stop annotations Bad assembly can affect ortho clusters: - amplifications or decreases of gene family numbers 19

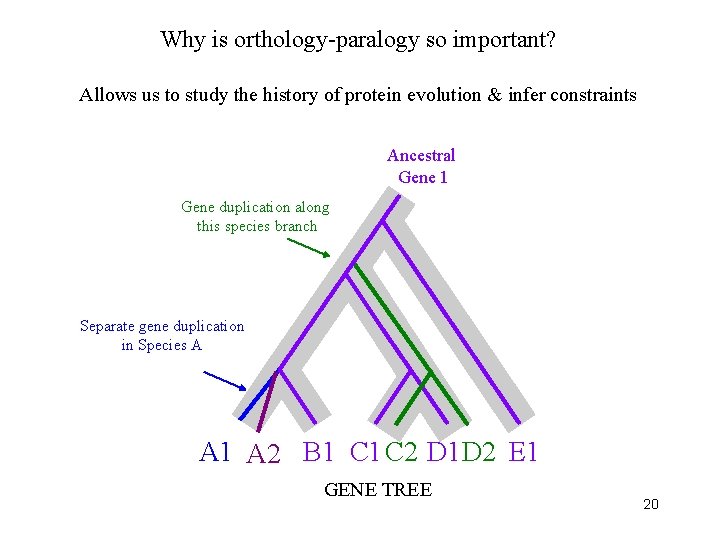
Why is orthology-paralogy so important? Allows us to study the history of protein evolution & infer constraints Ancestral Gene 1 Gene duplication along this species branch Separate gene duplication in Species A A 1 A 2 B 1 C 2 D 1 D 2 E 1 GENE TREE 20

21

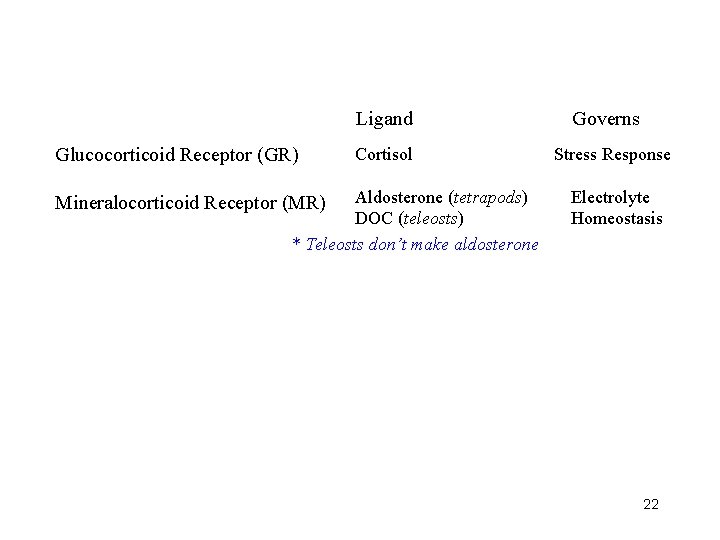
Ligand Glucocorticoid Receptor (GR) Cortisol Aldosterone (tetrapods) DOC (teleosts) * Teleosts don’t make aldosterone Mineralocorticoid Receptor (MR) Governs Stress Response Electrolyte Homeostasis 22

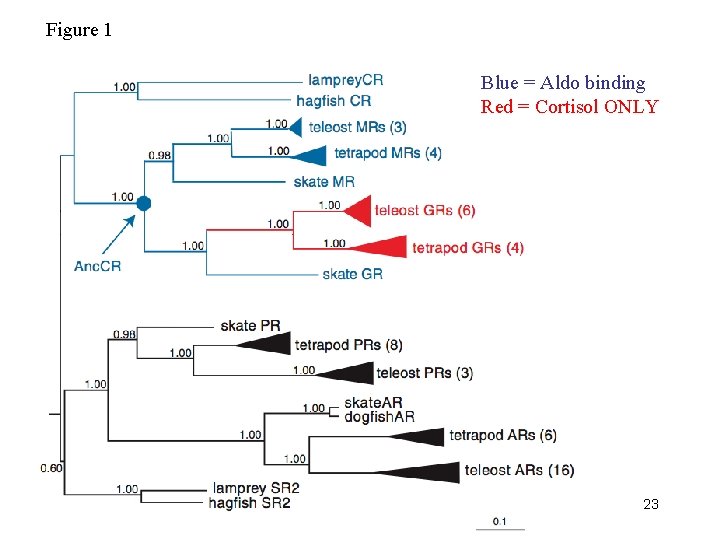
Figure 1 Blue = Aldo binding Red = Cortisol ONLY 23

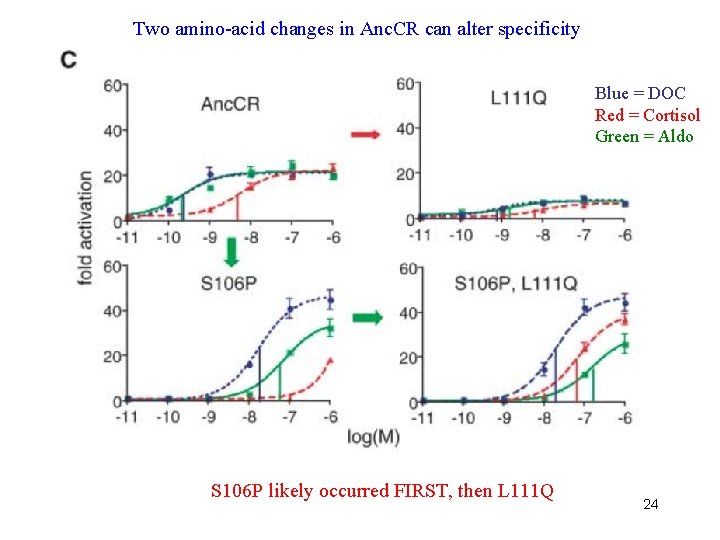
Two amino-acid changes in Anc. CR can alter specificity Blue = DOC Red = Cortisol Green = Aldo S 106 P likely occurred FIRST, then L 111 Q 24

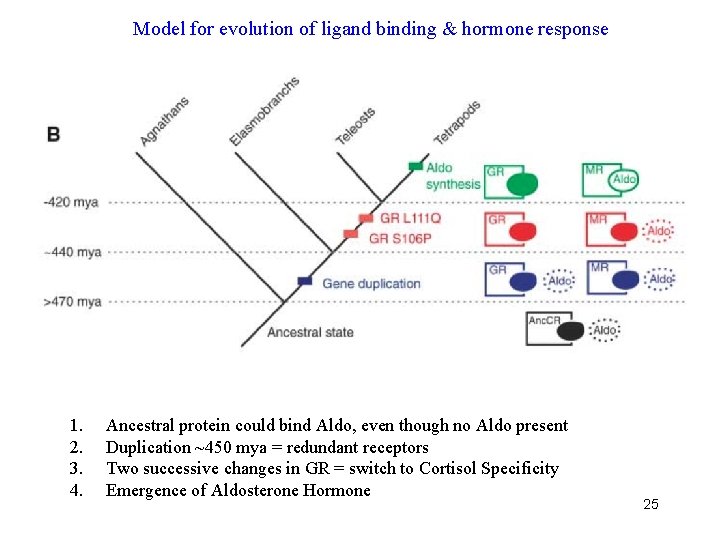
Model for evolution of ligand binding & hormone response 1. 2. 3. 4. Ancestral protein could bind Aldo, even though no Aldo present Duplication ~450 mya = redundant receptors Two successive changes in GR = switch to Cortisol Specificity Emergence of Aldosterone Hormone 25