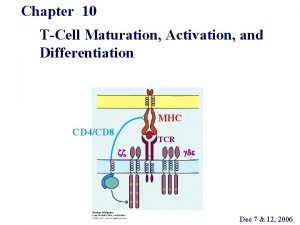
Organization of the MHC Organization of the MHC

















- Slides: 43

Organization of the MHC

Organization of the MHC The recent sequencing of the MHC as part of the genome project revealed numerous genes that function in antigen processing and other roles in the immune response, apart from the class I and class II MHC genes The MHC is a unique multigenic and polymorphic gene complex that regulates the adaptive immune response

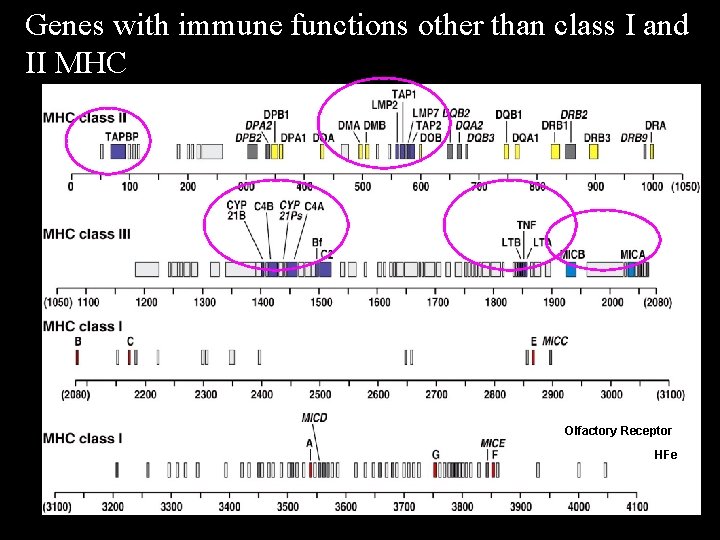
Genes with immune functions other than class I and II MHC Olfactory Receptor HFe

The MHC class I region contains additional class I genes other than the classic HLA-A, B and C MHC class IB genes (Non-classic) Innate Immunity • HLA-G the only MHC class I expressed on trophoblast and inhibits NK cell killing of fetus that would otherwise occur • HLA-E binds leader peptide of classic class I molecules, HLA-A, B and C and inhibits NK cell killing of target cell by engagement of inhibitory CD 94 / NKG 2 A receptor Limited polymorphism, restricted tissue expression

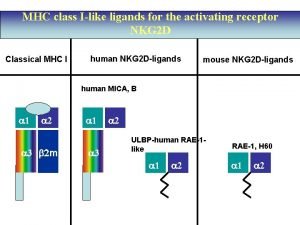
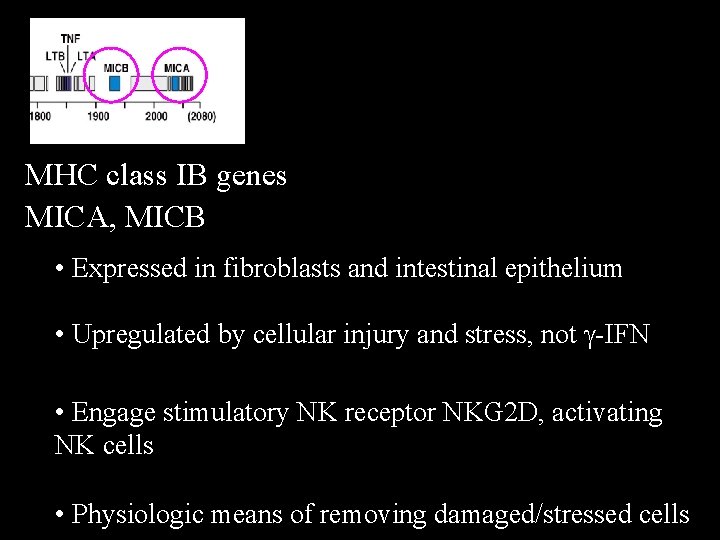
MHC class IB genes MICA, MICB • Expressed in fibroblasts and intestinal epithelium • Upregulated by cellular injury and stress, not g-IFN • Engage stimulatory NK receptor NKG 2 D, activating NK cells • Physiologic means of removing damaged/stressed cells

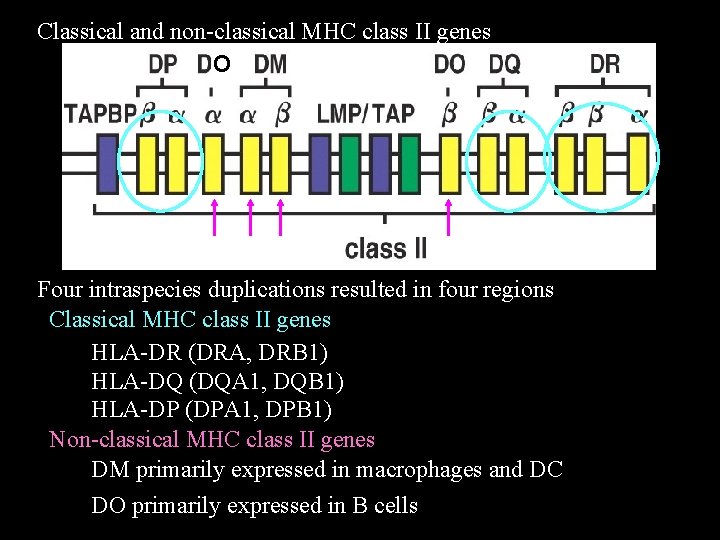
Classical and non-classical MHC class II genes O Four intraspecies duplications resulted in four regions Classical MHC class II genes HLA-DR (DRA, DRB 1) HLA-DQ (DQA 1, DQB 1) HLA-DP (DPA 1, DPB 1) Non-classical MHC class II genes DM primarily expressed in macrophages and DC DO primarily expressed in B cells

Class II

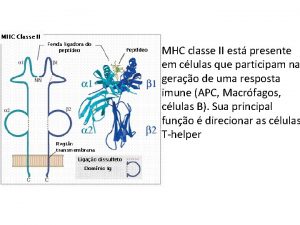
Distinctive features of peptide binding to class II MHC molecules Peptide binding groove open and no binding of N or C termini • Peptide antigen length variable: 12 -24 amino acids and length not critical • Pockets tether central portions of peptide and the pockets used vary considerably for loci and alleles Class II is a two chain molecule with both chains encoded in MHC and each contributes to peptide binding groove • Trans combinations of chains from different HLA-DQ or DP haplotypes generate additional post translational diversity • Polymorphism influences chain pairing and only certain gene pairs found=linkage disequilibrium

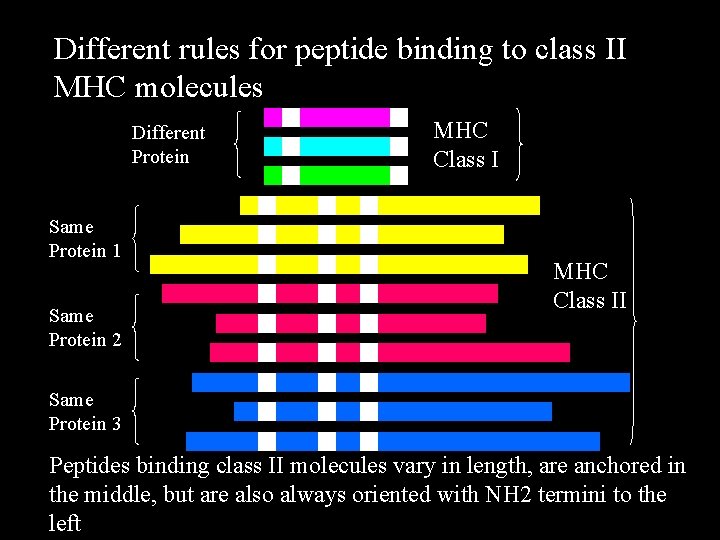
Different rules for peptide binding to class II MHC molecules Different Protein Same Protein 1 Same Protein 2 MHC Class II Same Protein 3 Peptides binding class II molecules vary in length, are anchored in the middle, but are also always oriented with NH 2 termini to the left

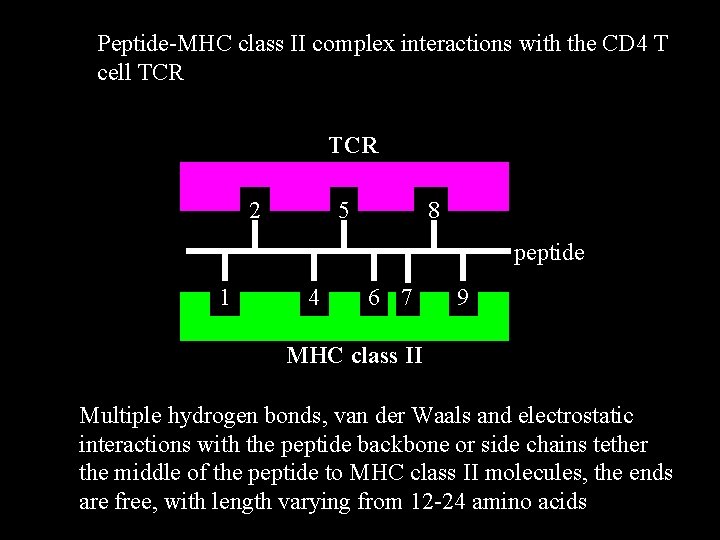
Peptide-MHC class II complex interactions with the CD 4 T cell TCR 2 5 8 peptide 1 4 6 7 9 MHC class II Multiple hydrogen bonds, van der Waals and electrostatic interactions with the peptide backbone or side chains tether the middle of the peptide to MHC class II molecules, the ends are free, with length varying from 12 -24 amino acids

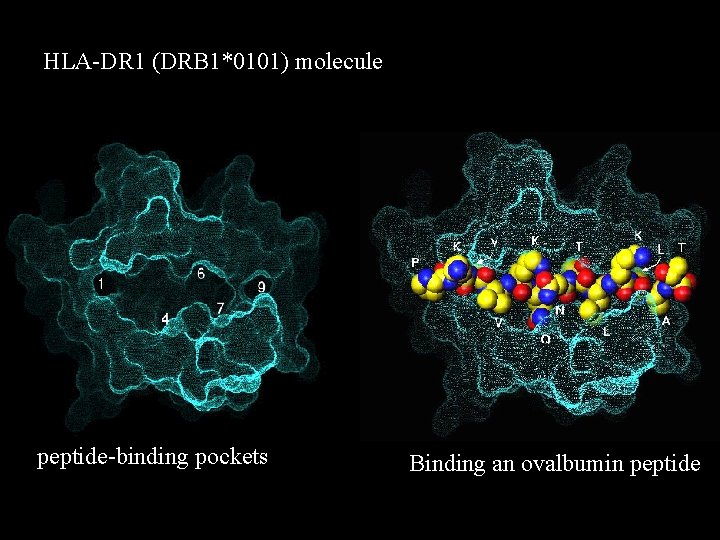
HLA-DR 1 (DRB 1*0101) molecule peptide-binding pockets Binding an ovalbumin peptide


MHC class II molecule binding a peptide P 7 P 2 Multiple bonds exist between peptide and MHC II P 8

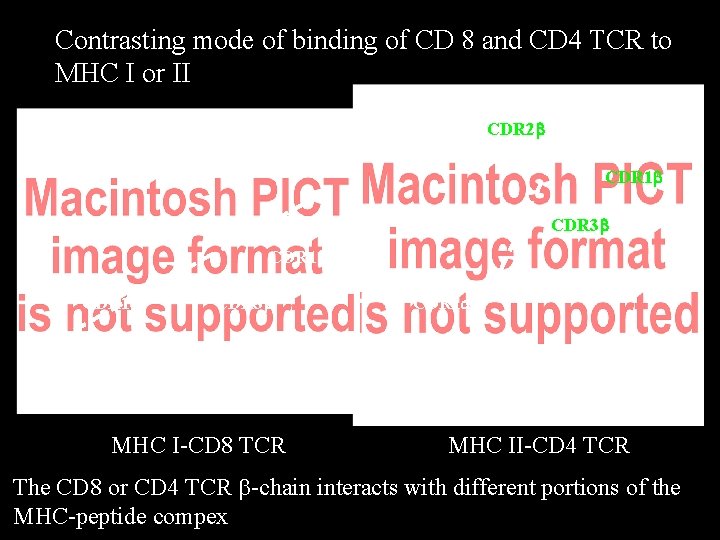
Contrasting mode of binding of CD 8 and CD 4 TCR to MHC I or II CDR 2 b CDR 1 b CDR 3 a CDR 3 b CDR 1 a CDR 3 b CDR 2 a MHC I-CD 8 TCR CDR 1 a CDR 2 a MHC II-CD 4 TCR The CD 8 or CD 4 TCR b-chain interacts with different portions of the MHC-peptide compex

MHC I-CD 8 TCR MHC II-CD 4 TCR

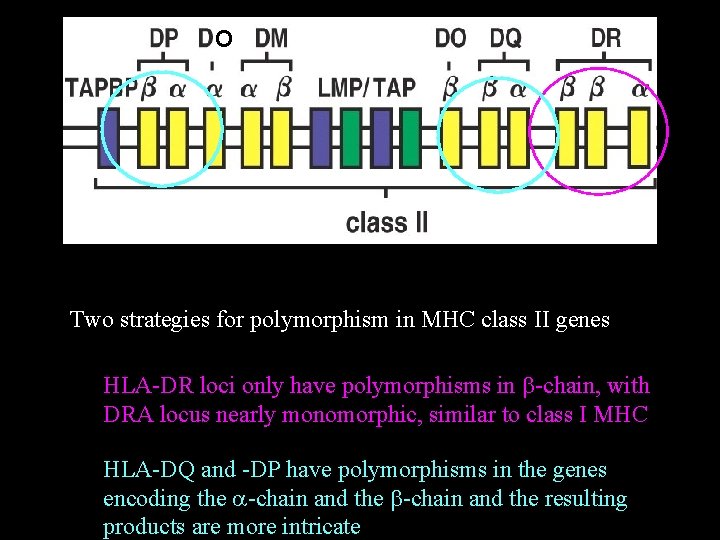
O Two strategies for polymorphism in MHC class II genes HLA-DR loci only have polymorphisms in b-chain, with DRA locus nearly monomorphic, similar to class I MHC HLA-DQ and -DP have polymorphisms in the genes encoding the a-chain and the b-chain and the resulting products are more intricate

HLA-DR loci only have polymorphisms in b-chain, with DRA locus nearly monomorphic, similar concept to class I MHC

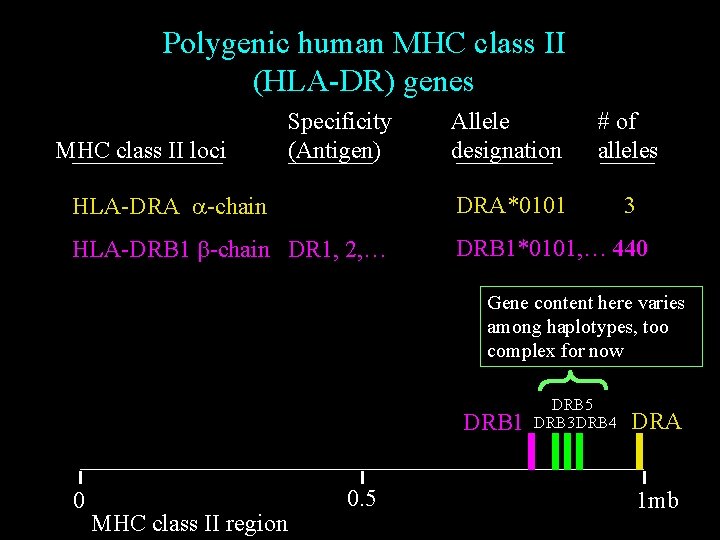
Polygenic human MHC class II (HLA-DR) genes Allele designation # of alleles HLA-DRA a-chain DRA*0101 3 HLA-DRB 1 b-chain DR 1, 2, … DRB 1*0101, … 440 MHC class II loci Specificity (Antigen) Gene content here varies among haplotypes, too complex for now DRB 1 0 MHC class II region 0. 5 DRB 3 DRB 4 DRA 1 mb

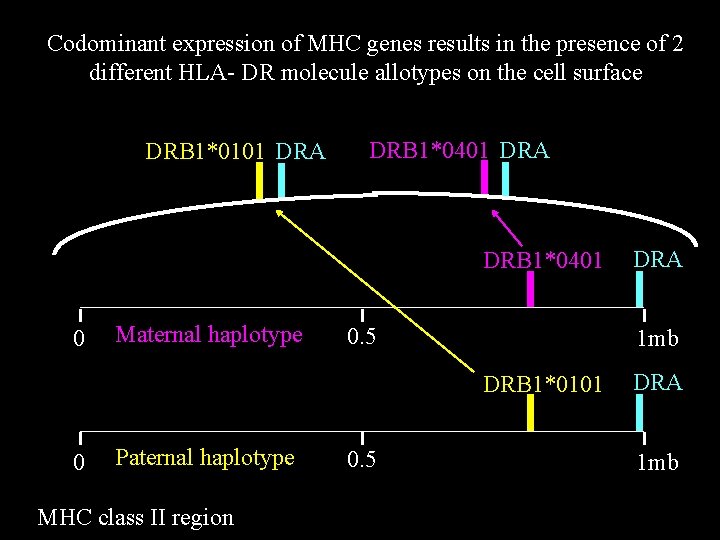
Codominant expression of MHC genes results in the presence of 2 different HLA- DR molecule allotypes on the cell surface DRB 1*0101 DRA DRB 1*0401 0 Maternal haplotype 0. 5 1 mb DRB 1*0101 0 Paternal haplotype MHC class II region 0. 5 DRA 1 mb

HLA-DR Polymorphisms NNN alleles of DRB 1 locus

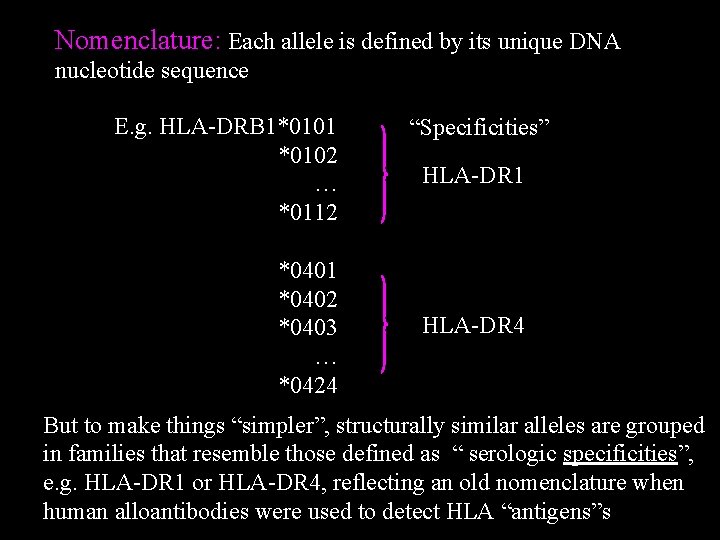
Nomenclature: Each allele is defined by its unique DNA nucleotide sequence E. g. HLA-DRB 1*0101 *0102 … *0112 *0401 *0402 *0403 … *0424 “Specificities” HLA-DR 1 HLA-DR 4 But to make things “simpler”, structurally similar alleles are grouped in families that resemble those defined as “ serologic specificities”, e. g. HLA-DR 1 or HLA-DR 4, reflecting an old nomenclature when human alloantibodies were used to detect HLA “antigens”s

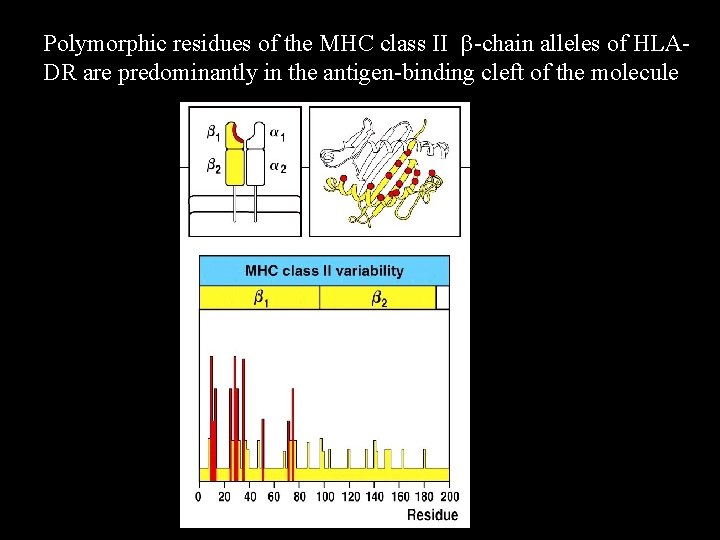
Polymorphic residues of the MHC class II b-chain alleles of HLADR are predominantly in the antigen-binding cleft of the molecule

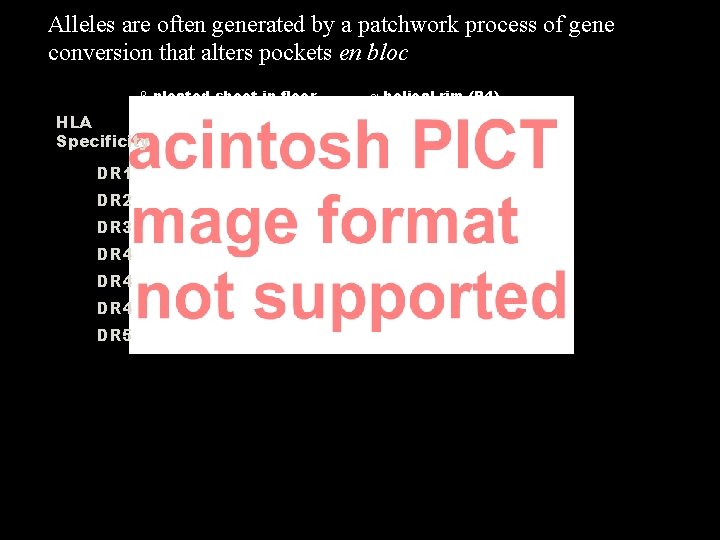
Alleles are often generated by a patchwork process of gene conversion that alters pockets en bloc b pleated sheet in floor HLA Specificity DR 1 DR 2 DR 3 DR 4 DR 5 a helical rim (P 4)

MHC class II molecule The most important difference among the HLA-DR 4 alleles is that their P 4 pockets differ 74 71 67 70 Amino acids forming floor of antigen groove Amino acids forming P 4 pocket in helical rim



What the actual sequence of some DRB 1 alleles looks like

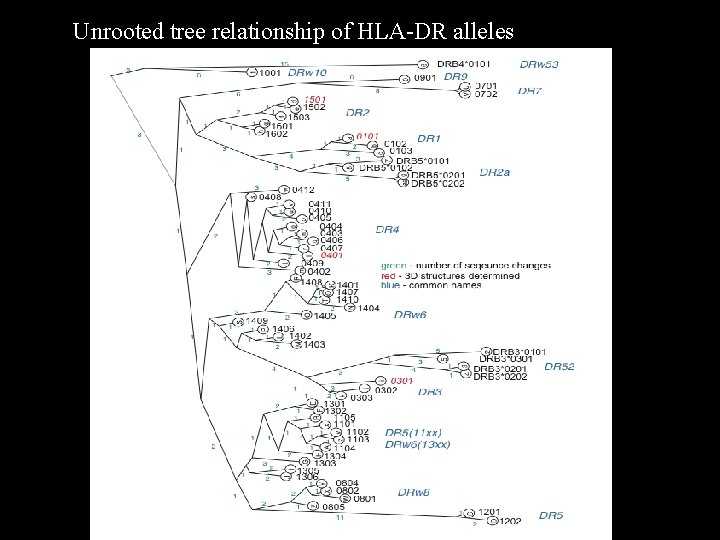
Unrooted tree relationship of HLA-DR alleles

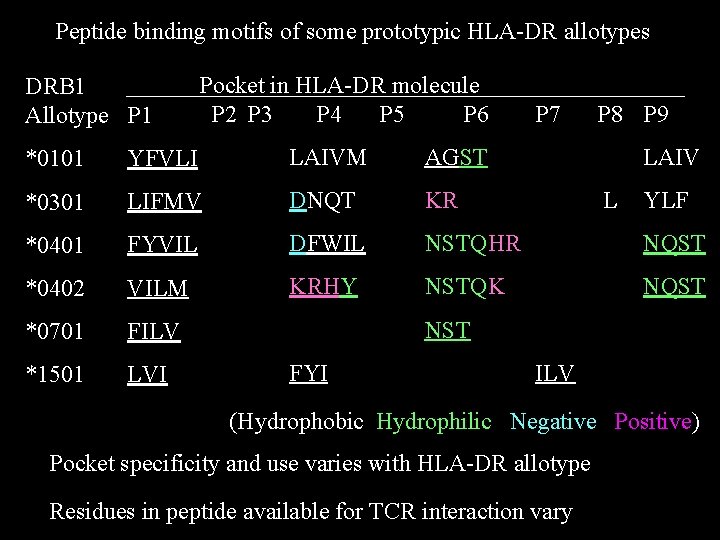
Peptide binding motifs of some prototypic HLA-DR allotypes DRB 1 Allotype P 1 Pocket in HLA-DR molecule P 2 P 3 P 4 P 5 P 6 P 7 P 8 P 9 *0101 YFVLI LAIVM AGST LAIV *0301 LIFMV DNQT KR *0401 FYVIL DFWIL NSTQHR NQST *0402 VILM KRHY NSTQK NQST *0701 FILV *1501 LVI L YLF NST FYI ILV (Hydrophobic Hydrophilic Negative Positive) Pocket specificity and use varies with HLA-DR allotype Residues in peptide available for TCR interaction vary

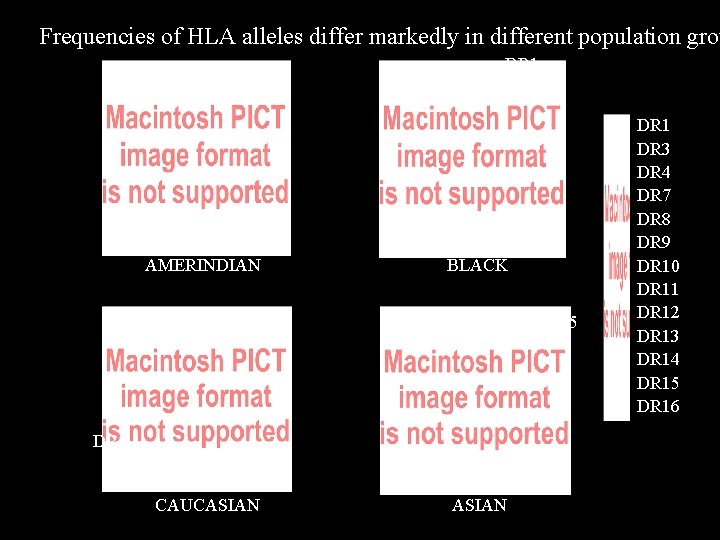
Frequencies of HLA alleles differ markedly in different population grou DR 1 DR 9 DR 4 AMERINDIAN BLACK DR 15 DR 7 CAUCASIAN DR 1 DR 3 DR 4 DR 7 DR 8 DR 9 DR 10 DR 11 DR 12 DR 13 DR 14 DR 15 DR 16

HLA-DQ

56 DQB 1 *0201 *0302 *0303 *0401 *0402 *0501 *0502 *0503 *0601 *0602 *0603 *0604 25 DQA 1 *0101 *0102 *0103 *0104 *0201 *0301 *0401 *0501 *0601 DRB 1 DRA Most common HLA-DQ alleles

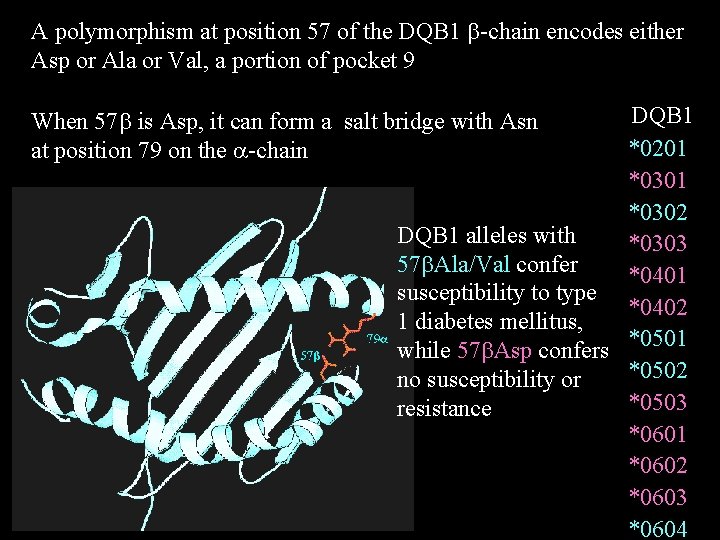
A polymorphism at position 57 of the DQB 1 b-chain encodes either Asp or Ala or Val, a portion of pocket 9 DQB 1 *0201 *0302 DQB 1 alleles with *0303 57 b. Ala/Val confer *0401 susceptibility to type *0402 1 diabetes mellitus, while 57 b. Asp confers *0501 *0502 no susceptibility or *0503 resistance *0601 *0602 *0603 *0604 When 57 b is Asp, it can form a salt bridge with Asn at position 79 on the a-chain

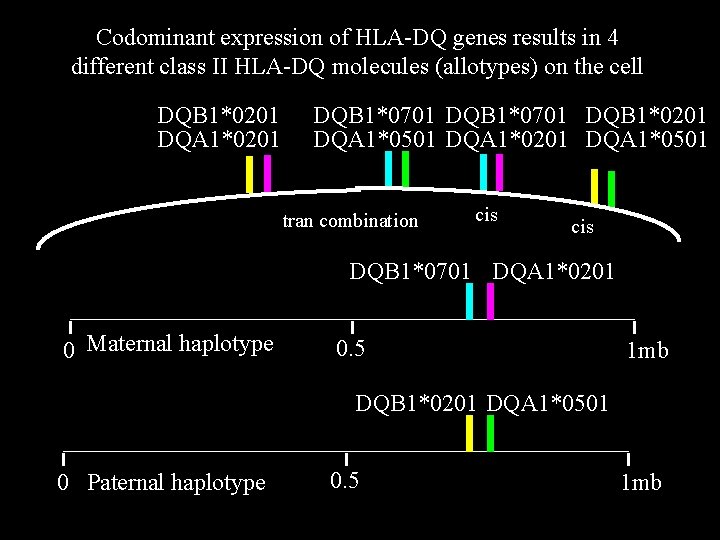
Codominant expression of HLA-DQ genes results in 4 different class II HLA-DQ molecules (allotypes) on the cell DQB 1*0201 DQA 1*0201 DQB 1*0701 DQB 1*0201 DQA 1*0501 DQA 1*0201 DQA 1*0501 tran combination cis DQB 1*0701 DQA 1*0201 0 Maternal haplotype 0. 5 1 mb DQB 1*0201 DQA 1*0501 0 Paternal haplotype 0. 5 1 mb

DQB 1 *0201 *0302 *0303 *0401 *0402 *0501 *0502 *0503 *0601 *0602 *0603 *0604 DQA 1 *0101 *0102 *0103 *0104 *0201 *0301 *0401 *0501 *0601 DRB 1 DRA Most common HLA-DQ alleles

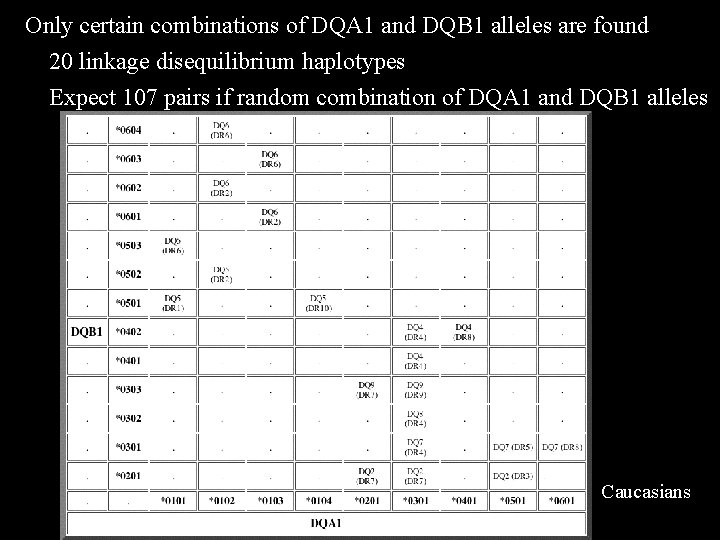
Only certain combinations of DQA 1 and DQB 1 alleles are found 20 linkage disequilibrium haplotypes Expect 107 pairs if random combination of DQA 1 and DQB 1 alleles Caucasians

Polymorphism of DQA 1 and DQB 1 influences chain pairing and only certain gene pairs are found together In some instances gene pairs not represented fail to produce an intact HLA-DQ molecule This linkage disequilibrium is centrally involved in determining the genetic basis of susceptibility to celiac disease and to type 1 diabetes mellitus The linkage disequilibrium extends throughout the class II region and sometimes across the entire MHC

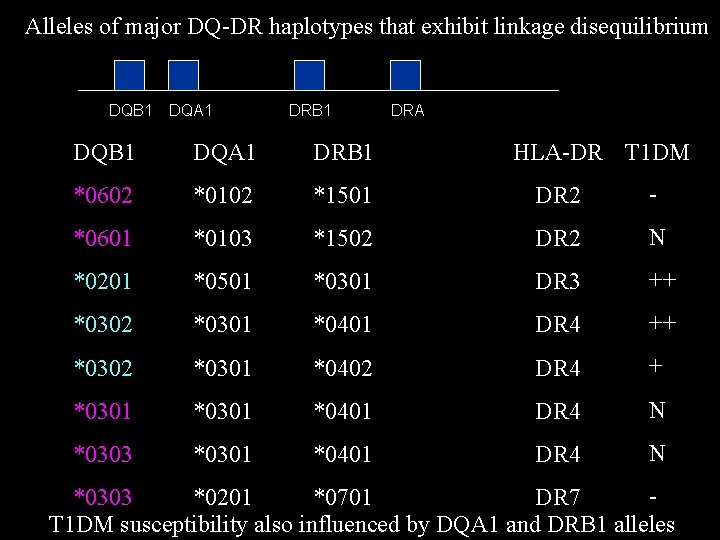
Alleles of major DQ-DR haplotypes that exhibit linkage disequilibrium DQB 1 DQA 1 DRB 1 DRA DQB 1 DQA 1 DRB 1 HLA-DR T 1 DM *0602 *0102 *1501 DR 2 - *0601 *0103 *1502 DR 2 N *0201 *0501 *0301 DR 3 ++ *0302 *0301 *0401 DR 4 ++ *0302 *0301 *0402 DR 4 + *0301 *0401 DR 4 N *0303 *0201 *0701 DR 7 T 1 DM susceptibility also influenced by DQA 1 and DRB 1 alleles


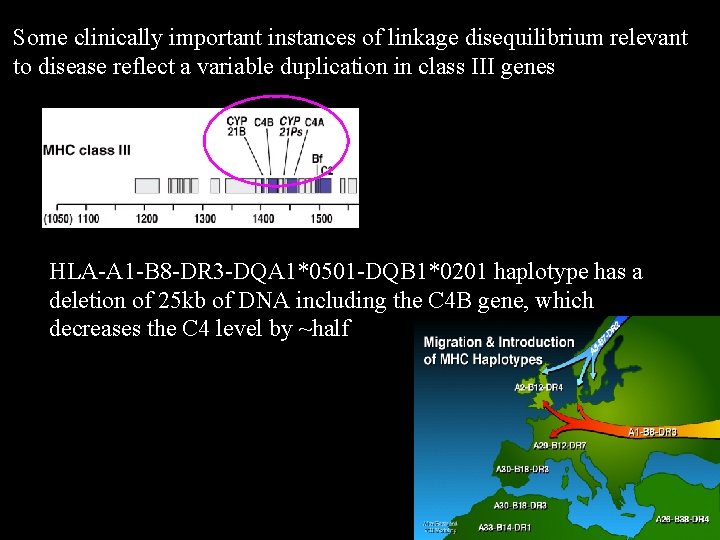
Some clinically important instances of linkage disequilibrium relevant to disease reflect a variable duplication in class III genes HLA-A 1 -B 8 -DR 3 -DQA 1*0501 -DQB 1*0201 haplotype has a deletion of 25 kb of DNA including the C 4 B gene, which decreases the C 4 level by ~half

Mispairing of class III regions that have one or two modules of C 4 during cross over can result in an additional loss of CYP 21 B(A 2) encodes steroid 21 hydroxylase and its absence results in congenital adrenal hyperplasia, a recessive disease due to a failure to synthesize cortisol; this results in androgenic precursor over- production Androgen excess leads to ambiguous genitalia in females, rapid somatic growth during childhood in both sexes with premature closure of the epiphyses and short adult stature (Infant Hercules)

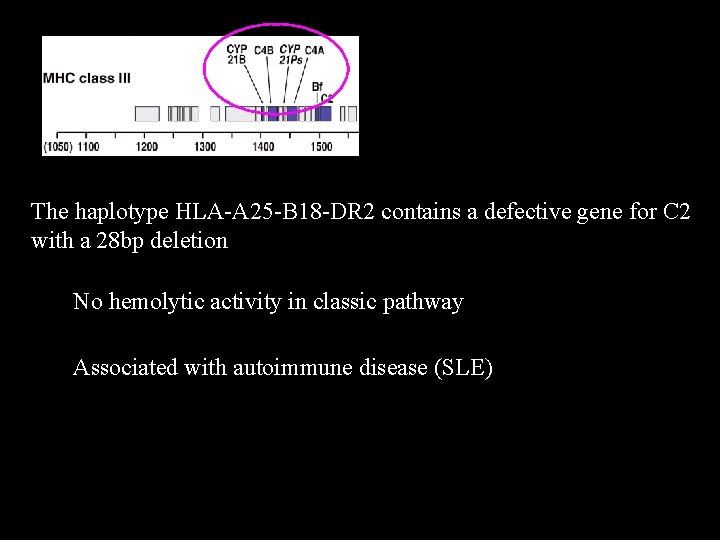
The haplotype HLA-A 25 -B 18 -DR 2 contains a defective gene for C 2 with a 28 bp deletion No hemolytic activity in classic pathway Associated with autoimmune disease (SLE)

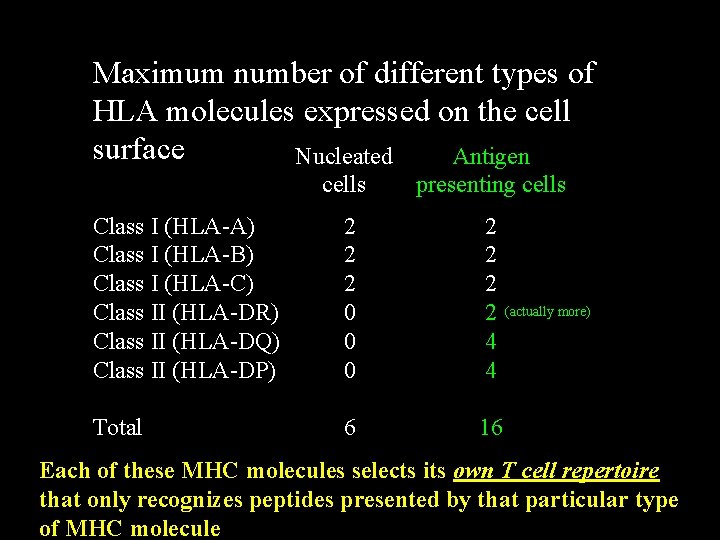
Maximum number of different types of HLA molecules expressed on the cell surface Nucleated Antigen cells presenting cells Class I (HLA-A) Class I (HLA-B) Class I (HLA-C) Class II (HLA-DR) Class II (HLA-DQ) Class II (HLA-DP) 2 2 2 0 0 0 2 2 4 4 Total 6 16 (actually more) Each of these MHC molecules selects its own T cell repertoire that only recognizes peptides presented by that particular type of MHC molecule
 Major histocompatibility complex structure
Major histocompatibility complex structure Apple health managed care plans
Apple health managed care plans Mhc molekula
Mhc molekula Mhc-pms
Mhc-pms Mhc software document self service
Mhc software document self service Mhc-pms in software engineering
Mhc-pms in software engineering Class i vs class ii mhc
Class i vs class ii mhc Mhc ii antigen presentation
Mhc ii antigen presentation Mhc and hla
Mhc and hla Mhc-pms meaning
Mhc-pms meaning Mhc-pms
Mhc-pms Mhc-pms use case diagram
Mhc-pms use case diagram Mhc-pms
Mhc-pms Ijvv de zwervers contributie
Ijvv de zwervers contributie Mhc ede
Mhc ede Eagle mhc
Eagle mhc Mhc-pms use case diagram
Mhc-pms use case diagram T cell mhc
T cell mhc Mhc molecules
Mhc molecules Células nucleadas
Células nucleadas Mhc pms in software engineering
Mhc pms in software engineering Homozygosty
Homozygosty T cell mhc
T cell mhc Mhc-pms meaning
Mhc-pms meaning Mhc evolution
Mhc evolution Mhc proteine
Mhc proteine Poligenia mhc
Poligenia mhc Process organization in computer organization
Process organization in computer organization Block organization example
Block organization example Môn thể thao bắt đầu bằng chữ đua
Môn thể thao bắt đầu bằng chữ đua Khi nào hổ con có thể sống độc lập
Khi nào hổ con có thể sống độc lập điện thế nghỉ
điện thế nghỉ Các loại đột biến cấu trúc nhiễm sắc thể
Các loại đột biến cấu trúc nhiễm sắc thể Nguyên nhân của sự mỏi cơ sinh 8
Nguyên nhân của sự mỏi cơ sinh 8 Trời xanh đây là của chúng ta thể thơ
Trời xanh đây là của chúng ta thể thơ Gấu đi như thế nào
Gấu đi như thế nào Thiếu nhi thế giới liên hoan
Thiếu nhi thế giới liên hoan Phối cảnh
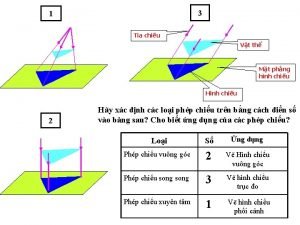
Phối cảnh Một số thể thơ truyền thống
Một số thể thơ truyền thống Các châu lục và đại dương trên thế giới
Các châu lục và đại dương trên thế giới Thế nào là hệ số cao nhất
Thế nào là hệ số cao nhất Hệ hô hấp
Hệ hô hấp Frameset trong html5
Frameset trong html5 Các số nguyên tố là gì
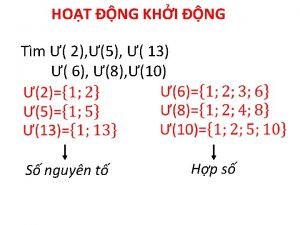
Các số nguyên tố là gì