Organization of the eukaryotic genomes Genome Size of








- Slides: 59

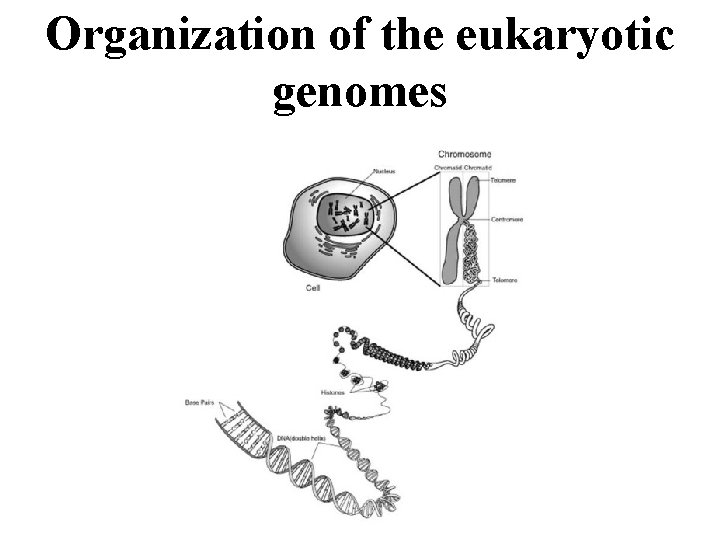
Organization of the eukaryotic genomes

Genome ØSize of genome? Nuclear / organelle genome ØDNA: coding, non-coding, repetitive DNA ØComplexity of genes v. Transposable elements v. Multigenes v. Pseudogenes Ø Regulatory sequences for Transcription? Ø Density of genes?

Genome organization • Prokaryotes – Most genome is coding – Small amount of non-coding is regulatory sequences • Eukaryotes – Most genome is non-coding (98%) – Regulatory sequences – Introns – Repetitive DNA

Prokaryote genomes • • Example: E. coli 89% coding 4, 285 genes 122 structural RNA genes Prophage remains Insertion sequence (IS) elements Horizontal transfers

Prokaryotic genome organization: • Haploid circular genomes (0. 5 -10 Mbp, 50010000 genes) • Operons: polycistronic transcription units • Environment-specific genes on plasmids and other types of mobile genetic elements • Usually asexual reproduction, great variety of recombination mechanisms • Transcription and translation take place in the same compartment

Eukaryotic genome • • • Example: C. elegans 10 chromosomes 19, 099 genes Coding region – 27% Average of 5 introns/gene Both long and short duplications

Eukaryotic genome organization 1. Multiple genomes: nuclear, plastid: mitochondria, chloroplasts 2. Plastid genomes resemble prokaryotic genomes 3. Multiple linear chromosomes, total size 510, 000 MB, 5000 to 50000 genes 4. Monocistronic transcription units 5. Discontinuous coding regions (introns and exons)

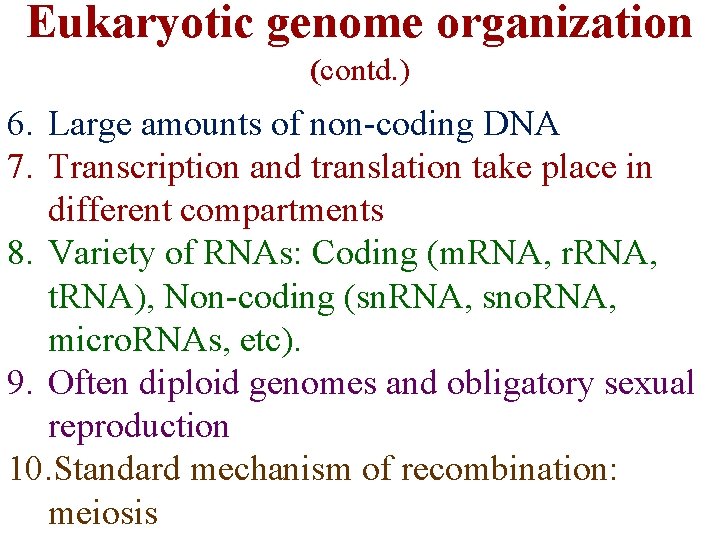
Eukaryotic genome organization (contd. ) 6. Large amounts of non-coding DNA 7. Transcription and translation take place in different compartments 8. Variety of RNAs: Coding (m. RNA, r. RNA, t. RNA), Non-coding (sn. RNA, sno. RNA, micro. RNAs, etc). 9. Often diploid genomes and obligatory sexual reproduction 10. Standard mechanism of recombination: meiosis

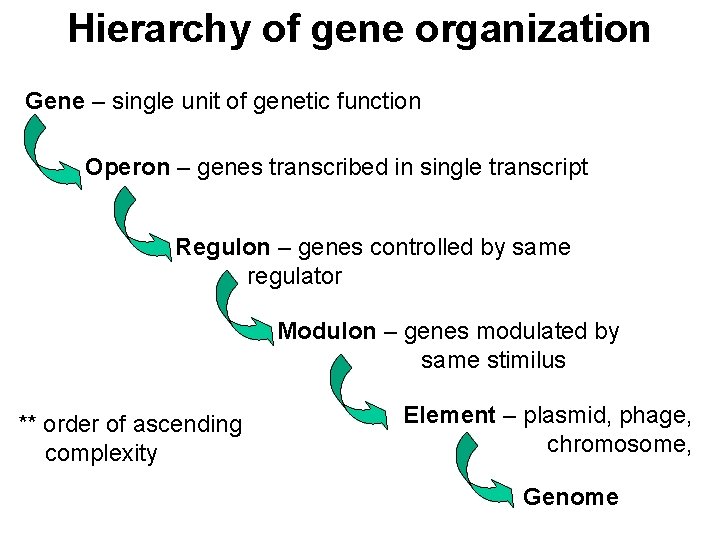
Hierarchy of gene organization Gene – single unit of genetic function Operon – genes transcribed in single transcript Regulon – genes controlled by same regulator Modulon – genes modulated by same stimilus ** order of ascending complexity Element – plasmid, phage, chromosome, Genome

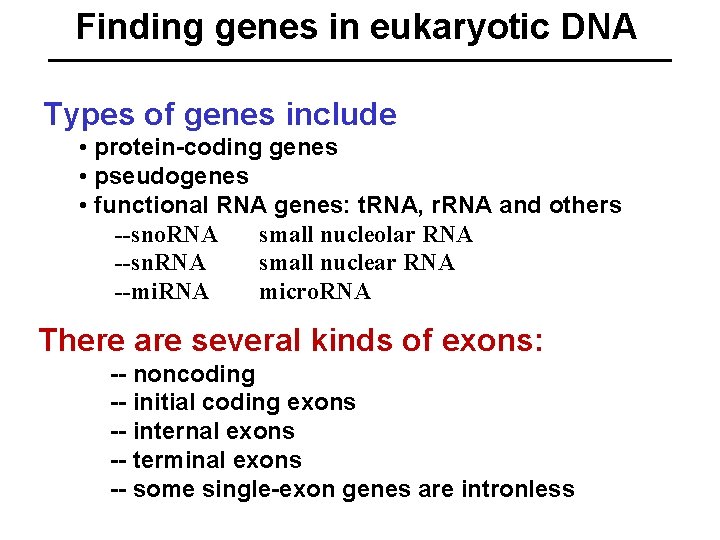
Finding genes in eukaryotic DNA Types of genes include • protein-coding genes • pseudogenes • functional RNA genes: t. RNA, r. RNA and others --sno. RNA small nucleolar RNA --sn. RNA small nuclear RNA --mi. RNA micro. RNA There are several kinds of exons: -- noncoding -- initial coding exons -- internal exons -- terminal exons -- some single-exon genes are intronless

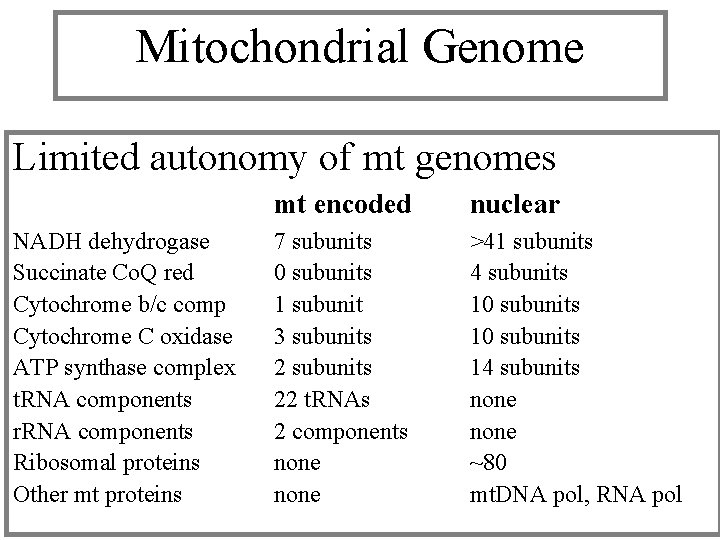
Mitochondrial Genome Limited autonomy of mt genomes NADH dehydrogase Succinate Co. Q red Cytochrome b/c comp Cytochrome C oxidase ATP synthase complex t. RNA components r. RNA components Ribosomal proteins Other mt proteins mt encoded nuclear 7 subunits 0 subunits 1 subunit 3 subunits 22 t. RNAs 2 components none >41 subunits 4 subunits 10 subunits 14 subunits none ~80 mt. DNA pol, RNA pol

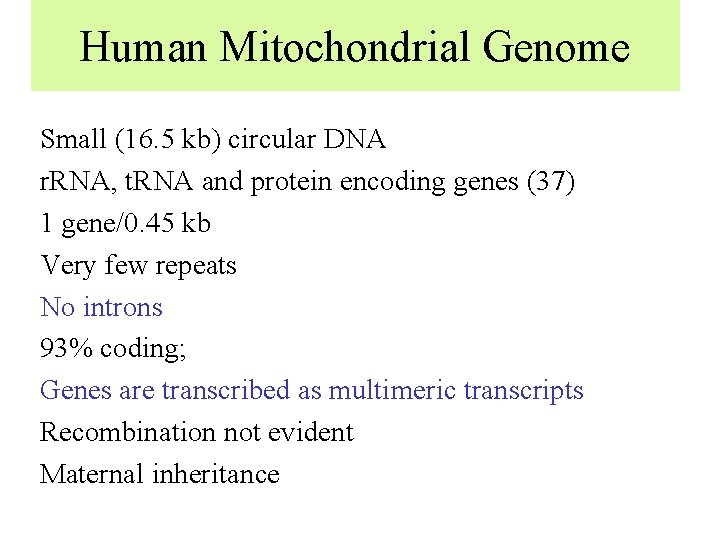
Human Mitochondrial Genome Small (16. 5 kb) circular DNA r. RNA, t. RNA and protein encoding genes (37) 1 gene/0. 45 kb Very few repeats No introns 93% coding; Genes are transcribed as multimeric transcripts Recombination not evident Maternal inheritance

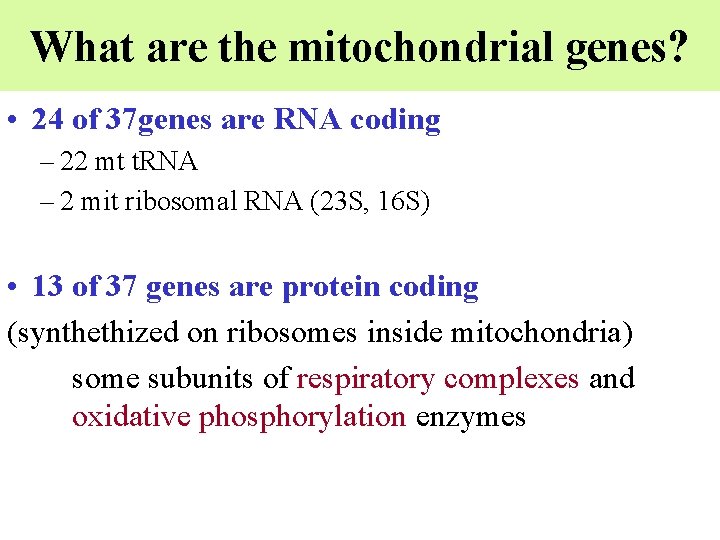
What are the mitochondrial genes? • 24 of 37 genes are RNA coding – 22 mt t. RNA – 2 mit ribosomal RNA (23 S, 16 S) • 13 of 37 genes are protein coding (synthethized on ribosomes inside mitochondria) some subunits of respiratory complexes and oxidative phosphorylation enzymes

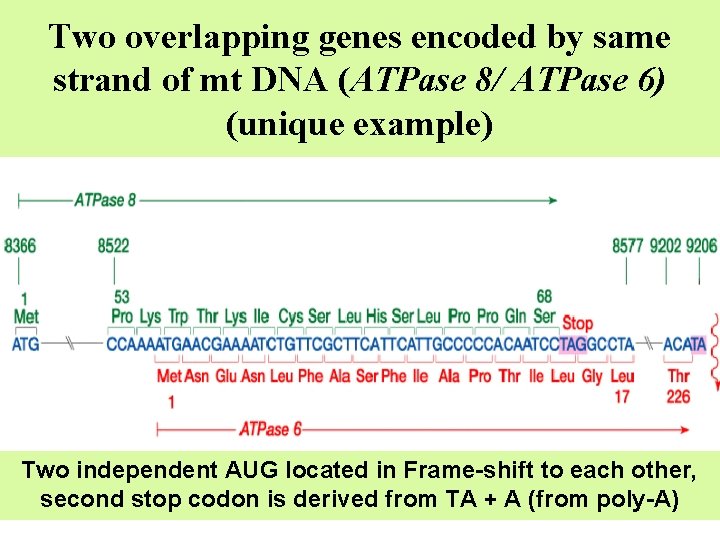
Two overlapping genes encoded by same strand of mt DNA (ATPase 8/ ATPase 6) (unique example) Two independent AUG located in Frame-shift to each other, second stop codon is derived from TA + A (from poly-A)

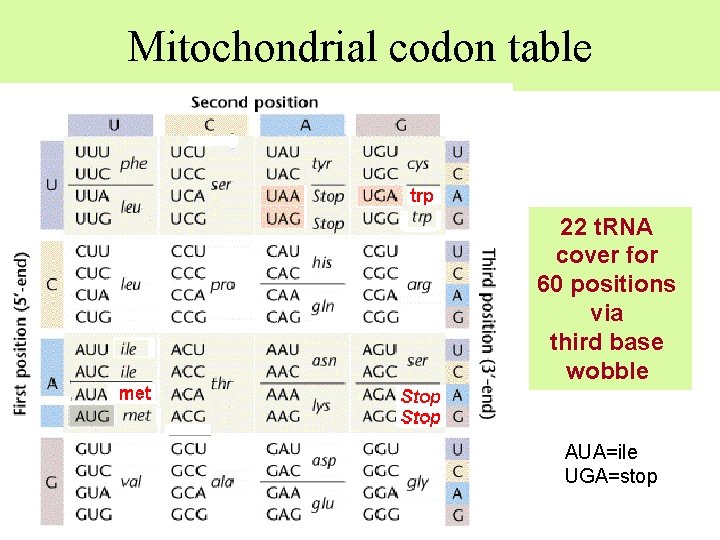
Mitochondrial codon table 22 t. RNA cover for 60 positions via third base wobble AUA=ile UGA=stop

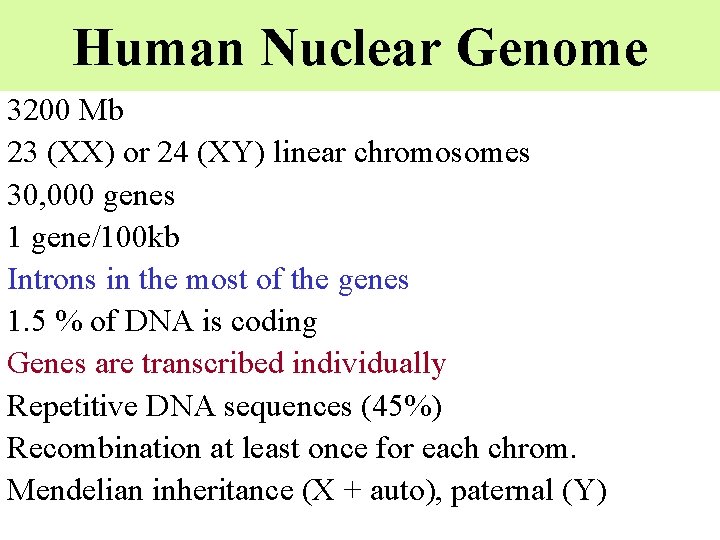
Human Nuclear Genome 3200 Mb 23 (XX) or 24 (XY) linear chromosomes 30, 000 genes 1 gene/100 kb Introns in the most of the genes 1. 5 % of DNA is coding Genes are transcribed individually Repetitive DNA sequences (45%) Recombination at least once for each chrom. Mendelian inheritance (X + auto), paternal (Y)

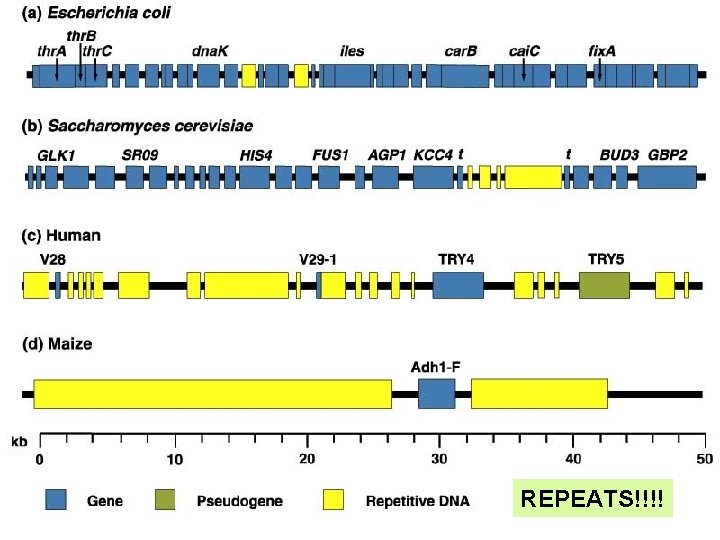
REPEATS!!!!

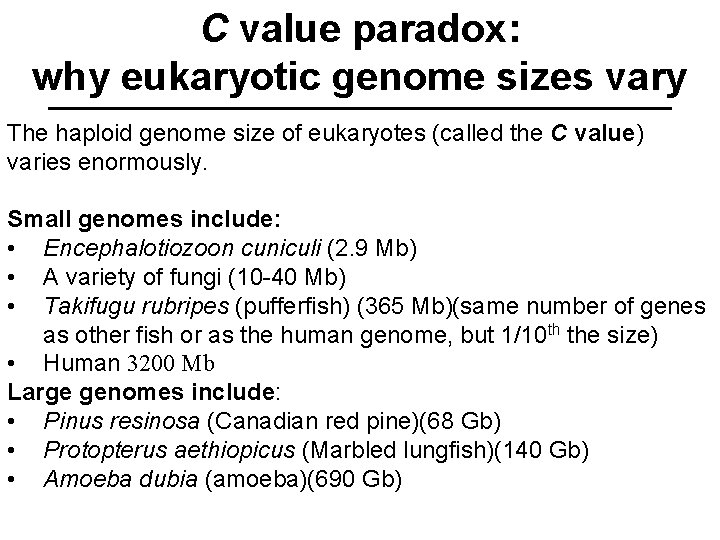
C value paradox: why eukaryotic genome sizes vary The haploid genome size of eukaryotes (called the C value) varies enormously. Small genomes include: • Encephalotiozoon cuniculi (2. 9 Mb) • A variety of fungi (10 -40 Mb) • Takifugu rubripes (pufferfish) (365 Mb)(same number of genes as other fish or as the human genome, but 1/10 th the size) • Human 3200 Mb Large genomes include: • Pinus resinosa (Canadian red pine)(68 Gb) • Protopterus aethiopicus (Marbled lungfish)(140 Gb) • Amoeba dubia (amoeba)(690 Gb)

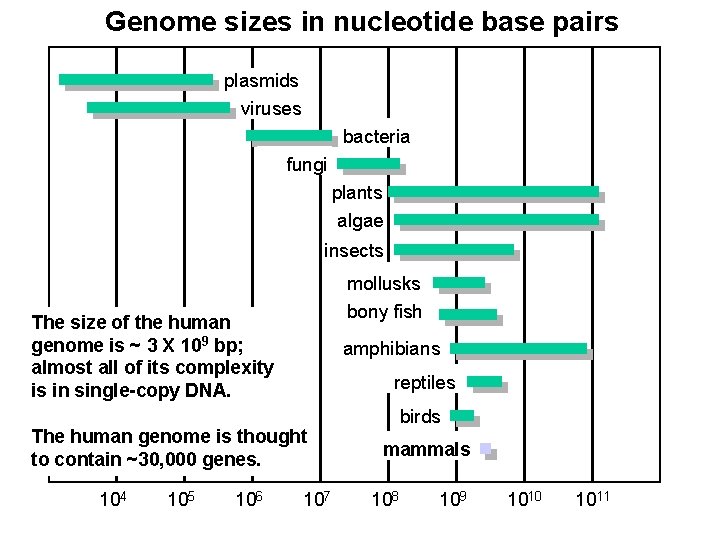
Genome sizes in nucleotide base pairs plasmids viruses bacteria fungi plants algae insects mollusks bony fish The size of the human genome is ~ 3 X 109 bp; almost all of its complexity is in single-copy DNA. amphibians reptiles The human genome is thought to contain ~30, 000 genes. 104 105 106 107 birds mammals 108 109 1010 1011

C value paradox: why eukaryotic genome sizes vary The range in C values does not correlate well with the complexity of the organism. This phenomenon is called the C value paradox. Why?

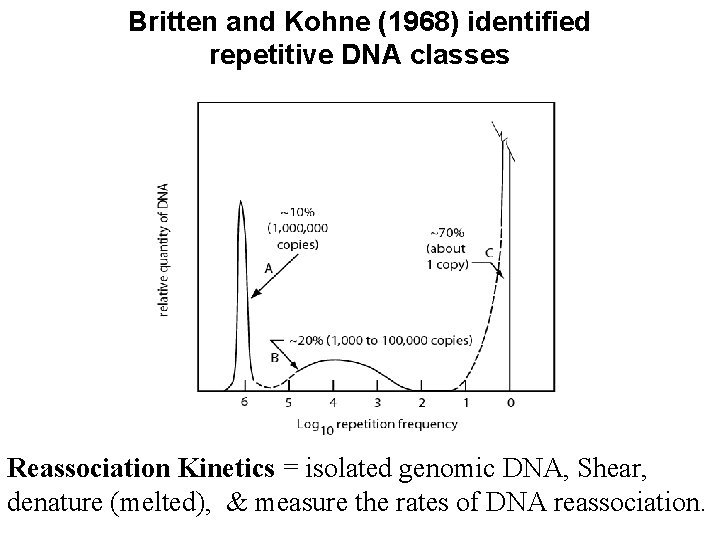
Britten and Kohne (1968) identified repetitive DNA classes Reassociation Kinetics = isolated genomic DNA, Shear, denature (melted), & measure the rates of DNA reassociation.

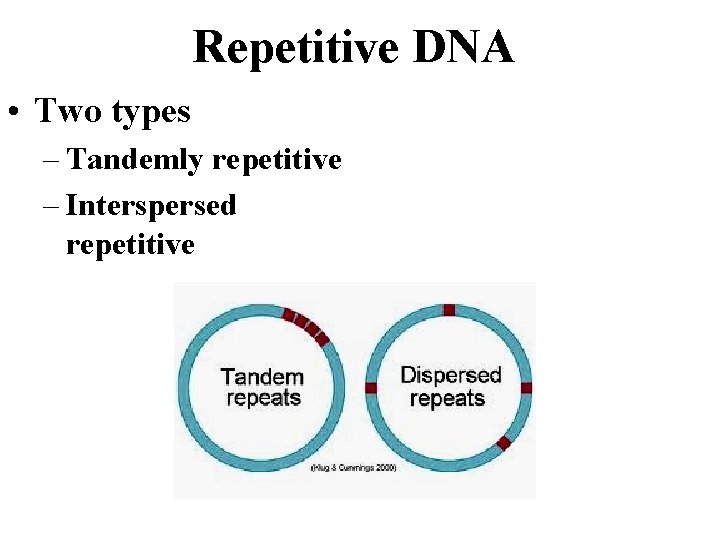
Repetitive DNA • Two types – Tandemly repetitive – Interspersed repetitive

Tandem repeats occur in DNA when a pattern of two or more nucleotides is repeated and the repetitions are adjacent to each other Form different density band on density gradient centrifugation (from bulk DNA) -satellite Example: A-T-T-C-G-A-T-T-C-G Tandem repeats: – Satellite DNA: – Microsatellite: – Minisatellite:

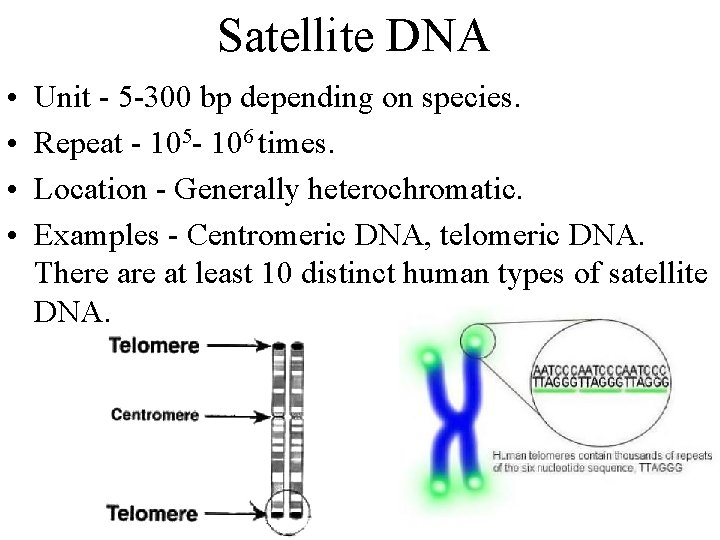
Satellite DNA • • Unit - 5 -300 bp depending on species. Repeat - 105 - 106 times. Location - Generally heterochromatic. Examples - Centromeric DNA, telomeric DNA. There at least 10 distinct human types of satellite DNA.

Microsatellite DNA • • Unit - 2 -4 bp (most 2). Repeat - on the order of 10 -100 times. Location - Generally euchromatic. Examples - Most useful marker for population level studies. .

Minisatellite DNA • • Unit - 15 -400 bp (average about 20). Repeat - Generally 20 -50 times (1000 -5000 bp long). Location - Generally euchromatic. Examples - DNA fingerprints. Tandemly repeated but often in dispersed clusters. Also called VNTR’s (variable number tandem repeats).

Tandemly Repetitive DNA Can Cause Diseases: • Fragile X Syndrome – “CGG” is repeated hundreds or even thousands of times creating a “fragile” site on the X chromosome. – It leads to mental retardation. • Huntington's Disease – “CAG” repeat causes a protein to have long stretches of the amino acid glutamine. – Leads to a neurological disorder that results in death

Interspersed Repetitive DNA • Interspersed repetitive DNA accounts for 25– 40 % of mammalian DNA. • They are scattered randomly throughout the genome. • The units are 100 – 1000 base pairs long. • Copies are similar but not identical to each other. • Interspersed repetitive genes are not stably integrated in the genome; they move from place to place. • They can sometimes mess up good genes

Interspersed Repetitive DNA These are: • Retrotransposons (class I transposable elements) (copy and paste), copy themselves to RNA and then back to DNA (using reverse transcriptase) to integrate into the genome. • Transposons (Class II TEs) (cut and paste) uses transposases to makes a staggered sticky cut.

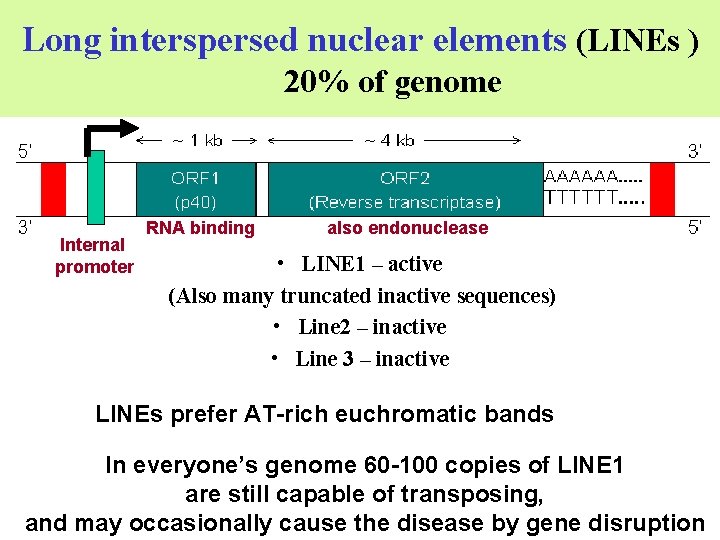
Interspersed Repetitive DNA • Retrotransposons are: v long terminal repeat (LTR) Any transposon flanked by Long Terminal Repeats. (also called retrovirus-like elements). None are active in humans, some are mobile in mice. v long interspersed nuclear elements (LINEs) encodes RT and v short interspersed nuclear elements (SINEs) uses RT from LINEs. example Alu made up of 350 base pairs long, recognized by the RE Alu. I (Non-autonomous)

Long interspersed nuclear elements (LINEs ) 20% of genome Internal promoter RNA binding also endonuclease • LINE 1 – active (Also many truncated inactive sequences) • Line 2 – inactive • Line 3 – inactive LINEs prefer AT-rich euchromatic bands In everyone’s genome 60 -100 copies of LINE 1 are still capable of transposing, and may occasionally cause the disease by gene disruption

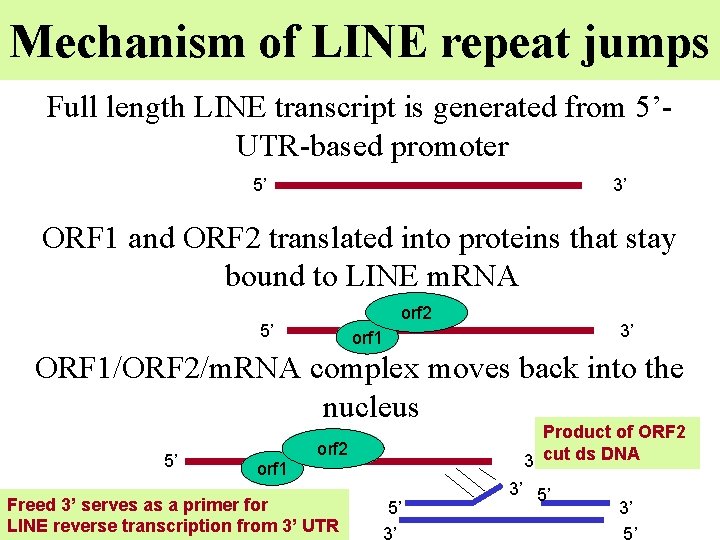
Mechanism of LINE repeat jumps Full length LINE transcript is generated from 5’UTR-based promoter 5’ 3’ ORF 1 and ORF 2 translated into proteins that stay bound to LINE m. RNA orf 2 5’ 3’ orf 1 ORF 1/ORF 2/m. RNA complex moves back into the nucleus 5’ orf 1 Product of ORF 2 3’ cut ds DNA orf 2 Freed 3’ serves as a primer for LINE reverse transcription from 3’ UTR 5’ 3’ 3’ 5’

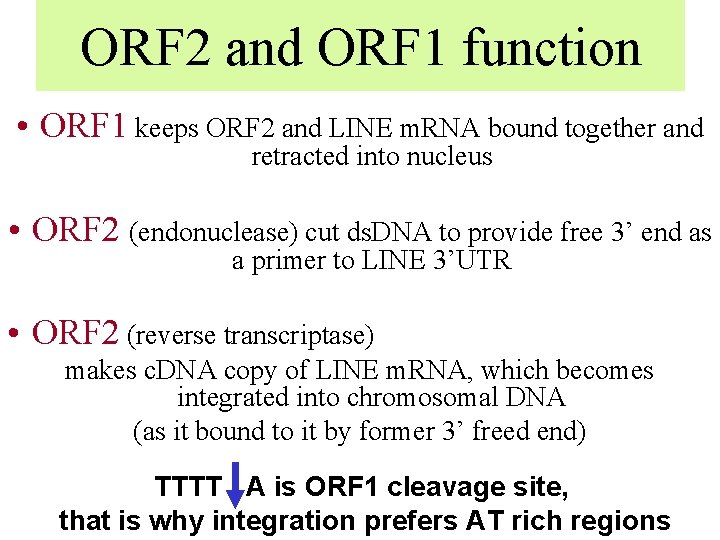
ORF 2 and ORF 1 function • ORF 1 keeps ORF 2 and LINE m. RNA bound together and retracted into nucleus • ORF 2 (endonuclease) cut ds. DNA to provide free 3’ end as a primer to LINE 3’UTR • ORF 2 (reverse transcriptase) makes c. DNA copy of LINE m. RNA, which becomes integrated into chromosomal DNA (as it bound to it by former 3’ freed end) TTTT A is ORF 1 cleavage site, that is why integration prefers AT rich regions

Short interspersed nuclear elements (SINE) 13% of genome • • Non-autonomous (no RT) 100 -400 bp long; No open reading frames (no start/stop codon) Derived from t. RNA (transcribed with RNA pol III, leaving internal promoter) • Depend on LINE machinery for its movement

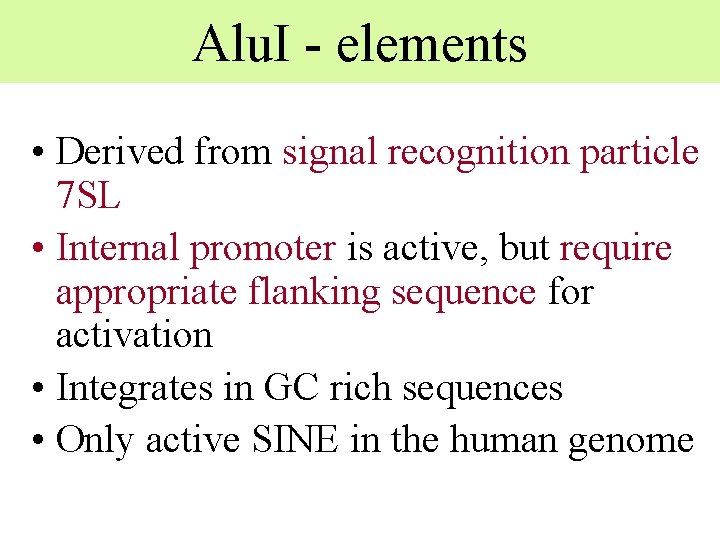
Alu. I - elements • Derived from signal recognition particle 7 SL • Internal promoter is active, but require appropriate flanking sequence for activation • Integrates in GC rich sequences • Only active SINE in the human genome

Diseases caused by Alu-integration • • Neurofibromatosis (Shwann cell tumors), haemophilia, breast cancer, Apert syndrome (distortions of the head and face and webbing of the hands and feet), cholinesterase deficiency (congenital myasthenic syndrome) complement deficiency (hereditary angioedema) α-thalassaemia Several types of cancer, including Ewing sarcoma, breast cancer, acute myelogenous leukaemia

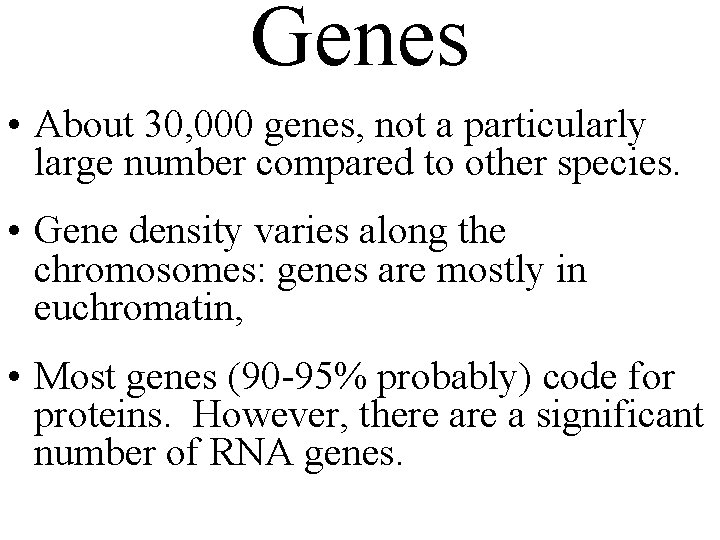
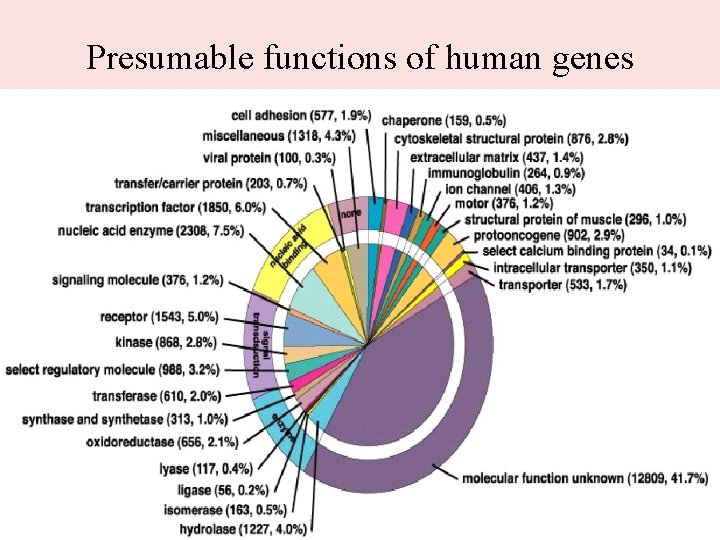
Genes • About 30, 000 genes, not a particularly large number compared to other species. • Gene density varies along the chromosomes: genes are mostly in euchromatin, • Most genes (90 -95% probably) code for proteins. However, there a significant number of RNA genes.

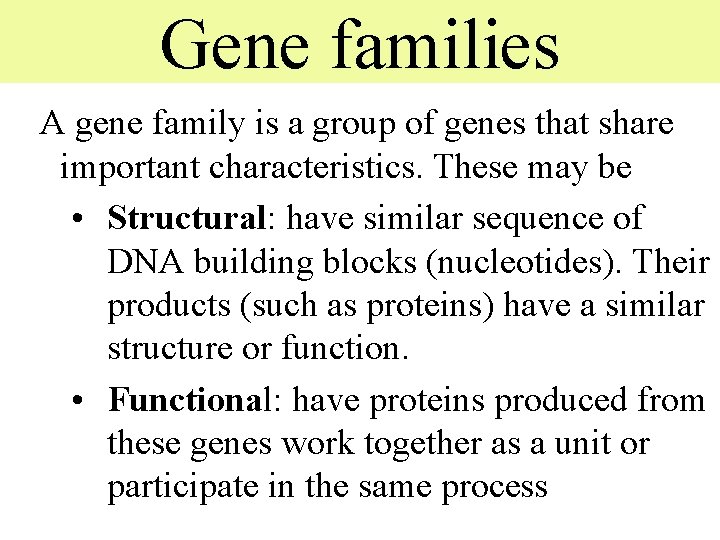
Gene families A gene family is a group of genes that share important characteristics. These may be • Structural: have similar sequence of DNA building blocks (nucleotides). Their products (such as proteins) have a similar structure or function. • Functional: have proteins produced from these genes work together as a unit or participate in the same process

Gene families (structural) 1. Classical gene families (overall conservativeness) Histones, alpha and betaglobines 2. Gene families with large conservative domains (other parts could be low conservative) HLH/b. ZIP box transcription factors 3. Gene families with short conservative motifs e. g. DEAD box (Asp-Glu-Ala-Asp), WD (Trp. Asp) repeat

Gene families (functional) 1 Regulatory protein gene families 2 Immune system proteins 3 Motor proteins 4 Signal transducing proteins 5 Transporters 6 Unclassified families

Multigene families Some genes are Transcribed (But Don't Make Proteins) • The entire family of genes probably evolved from a single ancestral gene. – Famous examples: r. RNA, globin genes – Four different pieces of r. RNA are used to make up a ribosome: 18 S, 5. 8 S, 28 S, and 5 S. – It turns out that three of these r. RNAs (18 S, 5. 8 S, 28 S, ) occur in the genome as a gene (on chrom 13, 14, 15, 21, 22) & transcribed together. (one 5 S on chrom. 1) – The entire multigene family is repeated nearly 300 times in clusters on five different chromosomes! • It makes sense to have many repeats of this multigene family because each cell needs many ribosomes for protein synthesis

Multigene family: r. RNA Genes • RNA polymerase I synthesizes 45 S which matures into 28 S, 18 S and 5. 8 S r. RNAs • RNA polymerase II synthesizes m. RNAs and most sn. RNA and micro. RNAs. • RNA polymerase III synthesizes t. RNAs, r. RNA 5 S and other small RNAs found in the nucleus and cytosol.

t. RNA genes (497 nuclear genes + 324 putative pseudogenes) • Humans have fewer t. RNA genes that the worm (584), but more than the fly (284); • Frog (Xenopus laevis) has thousands of t. RNA genes; • Number of t. RNA genes correlates with size of the oocytes; In large oocytes lots of protein needs to be sythesized simultaneously.

Fascinating world of RNAs coding & non-coding

Non-coding RNAs • t. RNA & r. RNA • 4. 5 S & 7 S RNA (Signal Recognition Particles) • sn. RNA – Pre-m. RNA splicing • sno. RNA – r. RNA modification • si. RNA – small interfering RNA • g. RNA – guide RNA in RNA editing • Telomerase RNA – primer for telomeric DNA synthesis • tm. RNA is a hybrid molecule, half t. RNA, half m. RNA • Xist: The X chromosome silencing is mediated by Xist – a 16, 000 nt long nc. RNA • sh. RNA (small heterochromatic RNAs ): expresses only one allele while other is silenced • LNA Locked Nucleic Acid • pi. RNA Piwi-interacting RNA

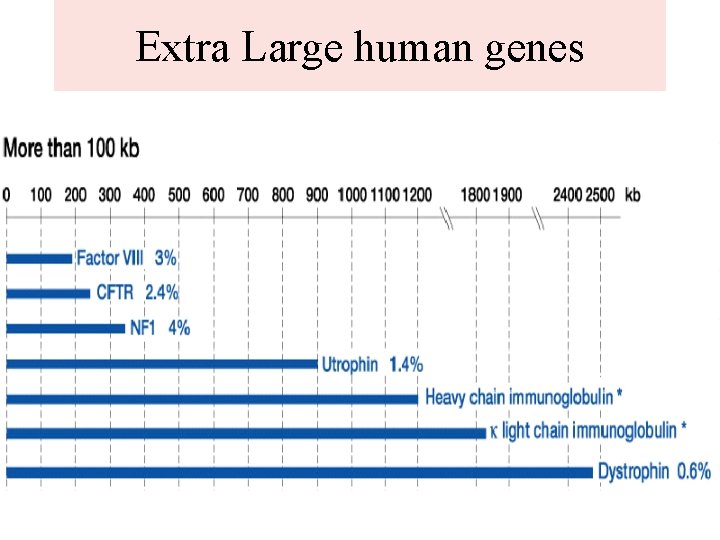
Protein-coding Genes • Genes vary greatly in size and organization. • Intron less: Some genes don’t have any introns. Most common example is the histone genes. • Some genes are quite huge: dystrophin (associated with Duchenne muscular dystrophy) is 2. 4 Mbp and takes 16 hours to transcribe. More than 99% of this gene is intron (total of 79 introns). • Highly expressed genes usually have short introns • Most exons are short: 200 bp on average. Intron size varies widely, from tens to millions of base pairs.

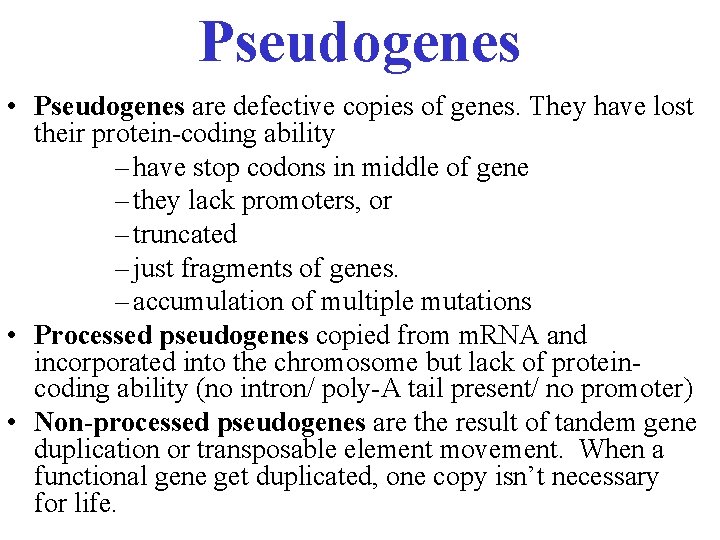
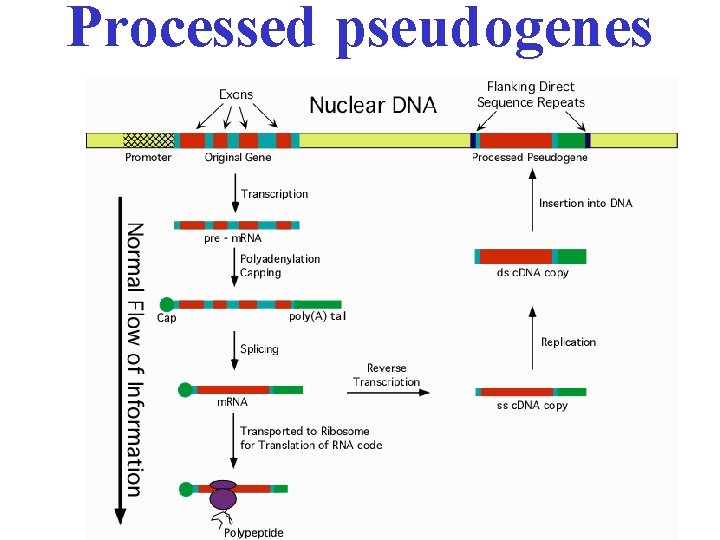
Pseudogenes • Pseudogenes are defective copies of genes. They have lost their protein-coding ability – have stop codons in middle of gene – they lack promoters, or – truncated – just fragments of genes. – accumulation of multiple mutations • Processed pseudogenes copied from m. RNA and incorporated into the chromosome but lack of proteincoding ability (no intron/ poly-A tail present/ no promoter) • Non-processed pseudogenes are the result of tandem gene duplication or transposable element movement. When a functional gene get duplicated, one copy isn’t necessary for life.

Processed pseudogenes

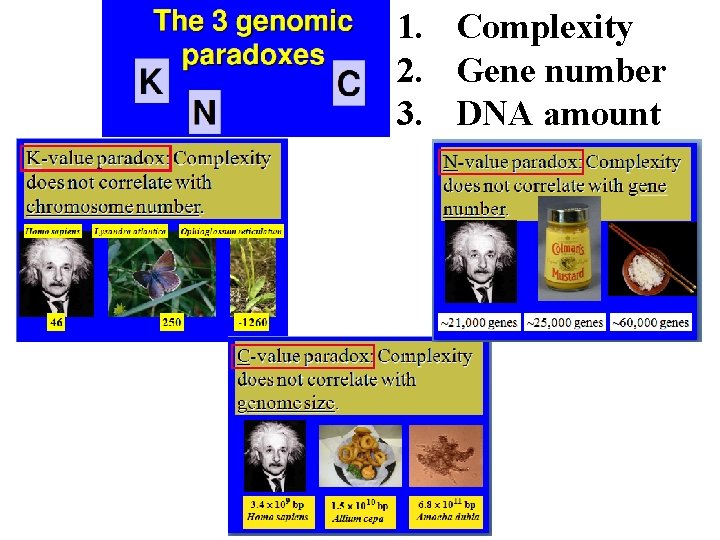
1. Complexity 2. Gene number 3. DNA amount

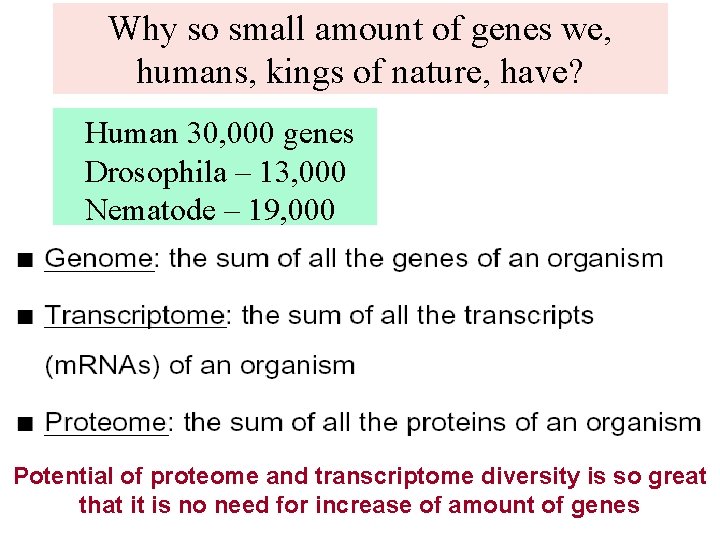
Why so small amount of genes we, humans, kings of nature, have? Human 30, 000 genes Drosophila – 13, 000 Nematode – 19, 000 Potential of proteome and transcriptome diversity is so great that it is no need for increase of amount of genes

Solutions ? Solution 1 to the N-value paradox: Many protein-encoding genes produce more than one protein product (e. g. , by alternative splicing or by RNA editing). Solution 2 to the N-value paradox: We are counting the wrong things, we should count other genetic elements (e. g. , small RNAs). Solution 3 to the N-value paradox: We should look at connectivity rather than at nodes. These should be exciting and should stimulate the next generation of genomic investigation. 51

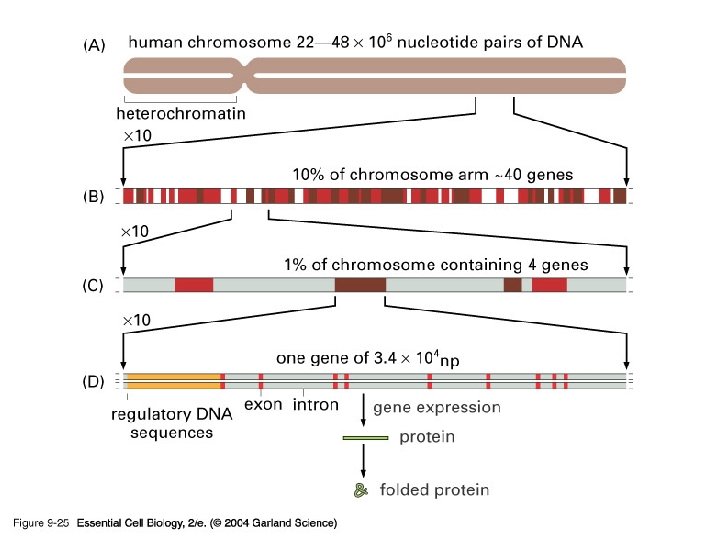
09_25_Chromosome 22. jpg

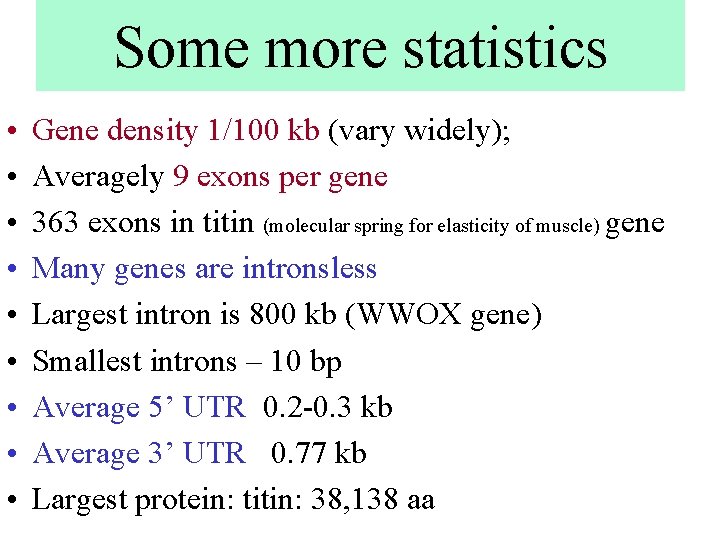
Some more statistics • • • Gene density 1/100 kb (vary widely); Averagely 9 exons per gene 363 exons in titin (molecular spring for elasticity of muscle) gene Many genes are intronsless Largest intron is 800 kb (WWOX gene) Smallest introns – 10 bp Average 5’ UTR 0. 2 -0. 3 kb Average 3’ UTR 0. 77 kb Largest protein: titin: 38, 138 aa

INTRONLESS GENES • • Interferon genes Histone genes Many ribonuclease genes Heat shock protein genes Many G-protein coupled receptors Some genes with HMG boxes Various neurotransmitters receptors and hormone receptors

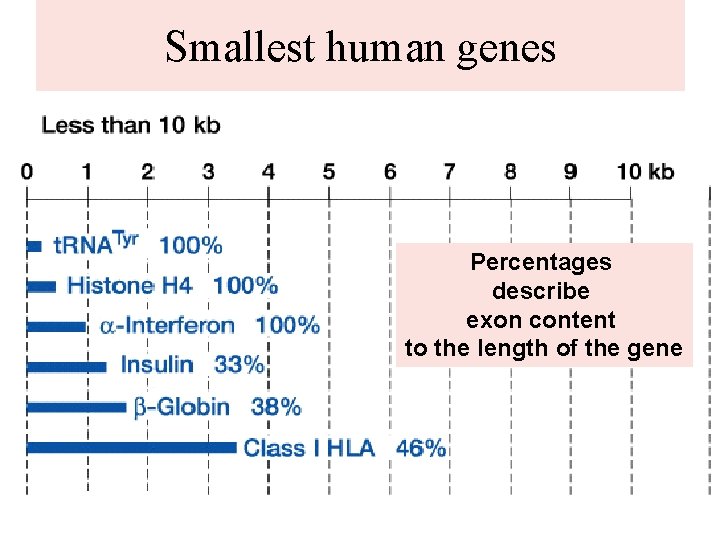
Smallest human genes Percentages describe exon content to the length of the gene

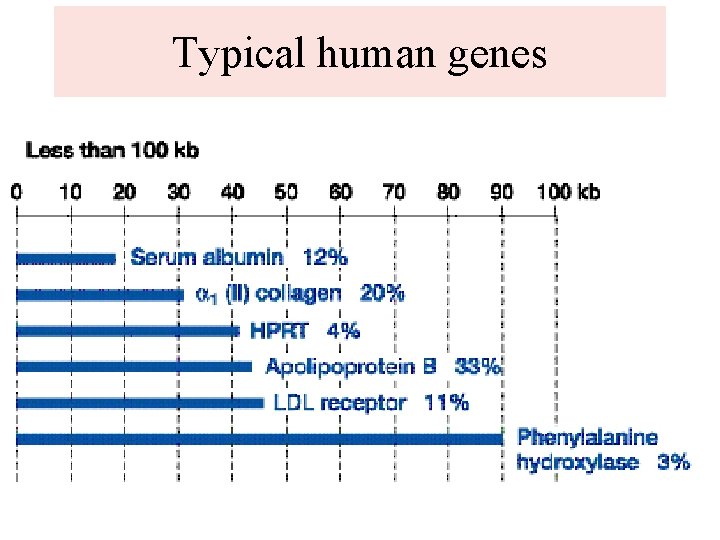
Typical human genes

Extra Large human genes

Presumable functions of human genes

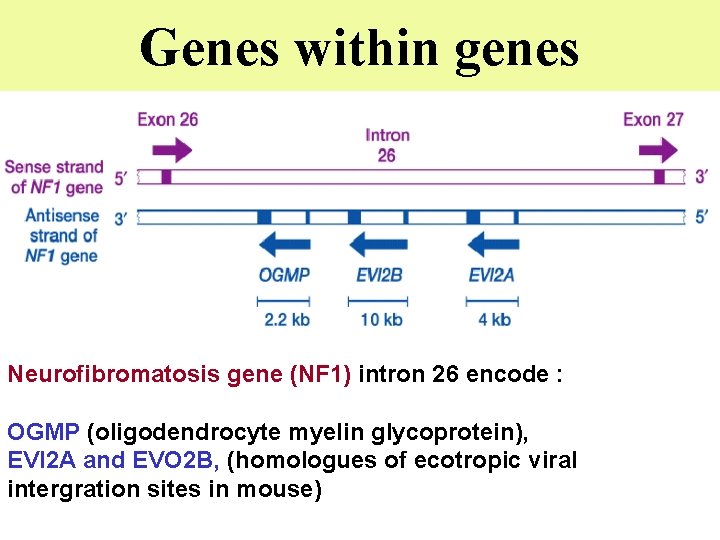
Genes within genes Neurofibromatosis gene (NF 1) intron 26 encode : OGMP (oligodendrocyte myelin glycoprotein), EVI 2 A and EVO 2 B, (homologues of ecotropic viral intergration sites in mouse)