ORF Calling ORF Calling Why Need to know

































- Slides: 54

ORF Calling

ORF Calling Why? Need to know protein sequence Protein sequence is usually what does the work Functional studies Crystallography Proteomics Similarity studies Proteins are better for remote similarities than DNA sequences Protein sequences change slower than DNA sequences

ORF Calling Extrinsic gene calling Compare your DNA sequences to known sequences. Needs other sequences that are known! Intrinsic gene calling Only use information in your DNA sequences. Does not use other information.

Extrinsic gene calling Start with DNA sequence Translate in all 6 reading frames

Why are there 6 reading frames? 3 2 1 AG TAA AAC TTT AAT TGG TTA A A GTA AAA CTT TAA TTG GTT AA AGT AAA ACT TTA ATT GTT GGT TAA ||| ||| TCA TTT TGA AAT TAA CCA ATT -1 -2 -3 TCA TTT TGA AAT TAA CCA ATT TC ATT TTG AAA TTA ACC AAT T T CAT TTT GAA ATT AAC CAA TT

Extrinsic gene calling Start with DNA sequence Translate in all 6 reading frames Compare your sequence to known protein sequences Find the ends of each, and call those genes!

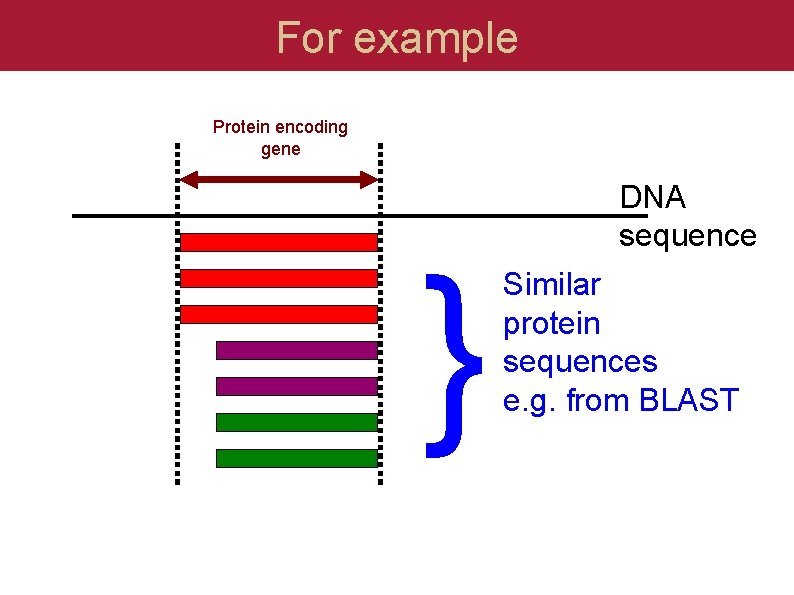
For example Protein encoding gene } DNA sequence Similar protein sequences e. g. from BLAST

Uses of extrinsic calling This is how (most) metagenome ORF calling is done Eukaryotic ORF calling – especially using EST sequences

Problems with extrinsic calling Very slow (depending on search algorithm) Dependent on your database Only finds known genes

Alternatives to extrinsic gene calling Intrinsic gene calling Ab initio gene calling What are the start codons? What are the stop codons? ATG TAA TAG TGA

How frequently do stop codons appear? Approximately once every 20 amino acids at random! A stretch of 100 amino acids is likely to have a stop codon!

How to call ORFs (the easy way) 3 2 1 DNA -1 -2 -3

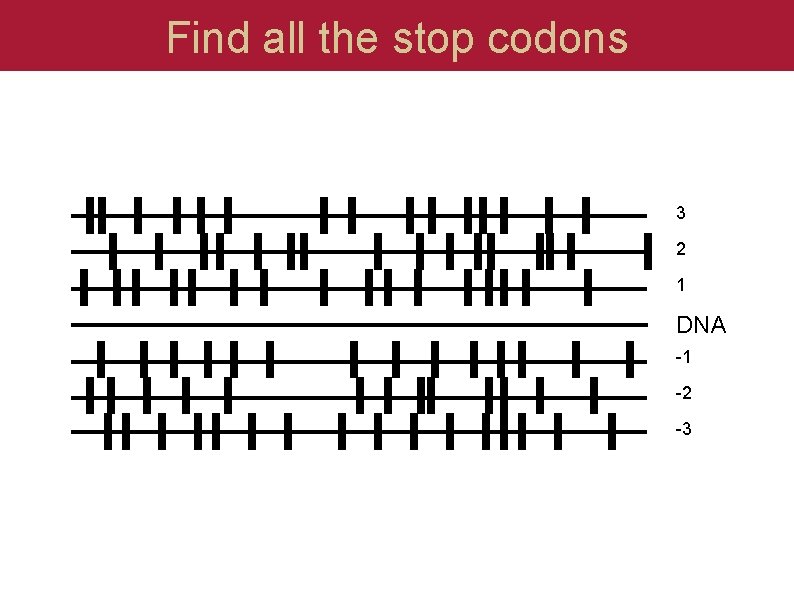
Find all the stop codons 3 2 1 DNA -1 -2 -3

Find all the ORFs > x amino acids X is often 100 amino acids 3 2 1 DNA -1 -2 -3

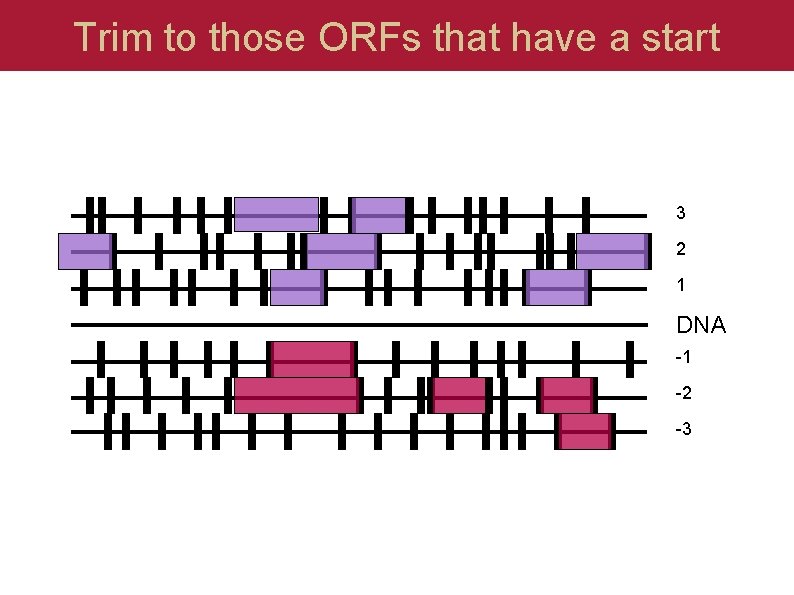
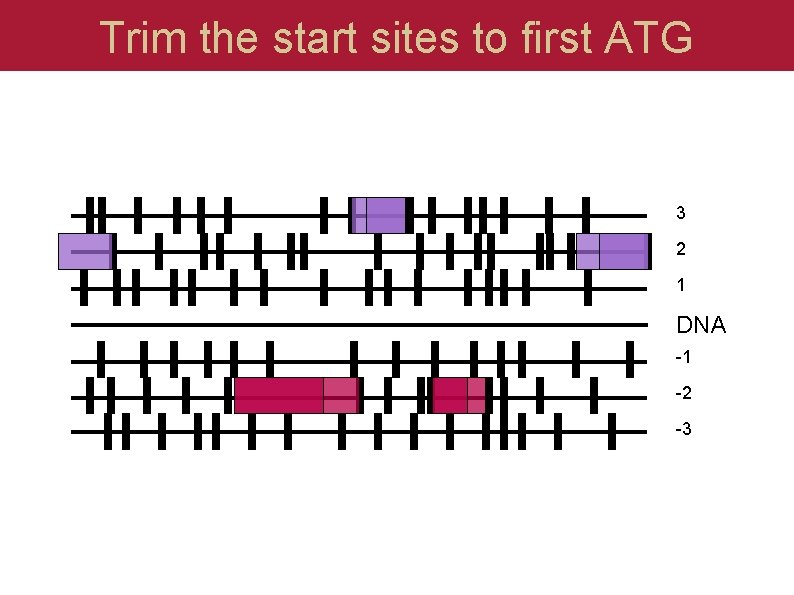
Trim to those ORFs that have a start 3 2 1 DNA -1 -2 -3

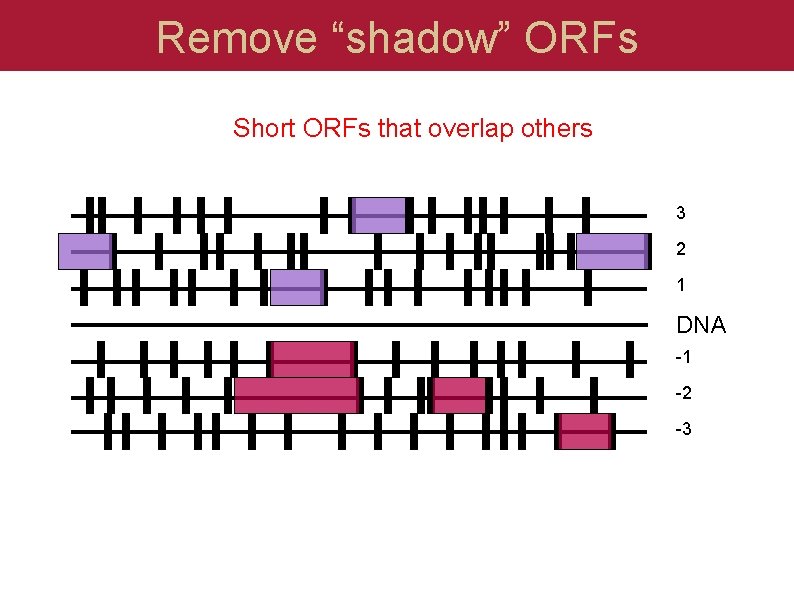
Remove “shadow” ORFs Short ORFs that overlap others 3 2 1 DNA -1 -2 -3

Trim the start sites to first ATG 3 2 1 DNA -1 -2 -3

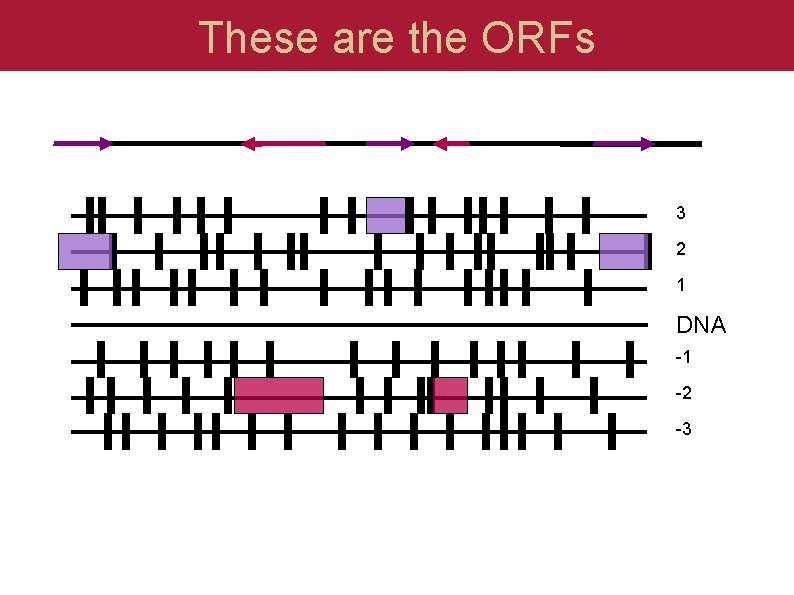
These are the ORFs 3 2 1 DNA -1 -2 -3

Intrinsic ORF calling using Markov Models

Markov Models Based on language processing Common for gene and protein finding, alignments, and so on

What is the most common word? English: the Spanish: el (la) Portuguese: que

Scrabble

Scrabble In scrabble, how do they score the letters? The most abundant letters (easiest to place on the board) are given the lowest score

Scrabble 1 point: E, A, I, O, N, R, T, L, S, U 2 points: D, G 3 points: B, C, M, P 4 points: F, H, V, W, Y 5 points: K 8 points: J, X 10 points: Q, Z

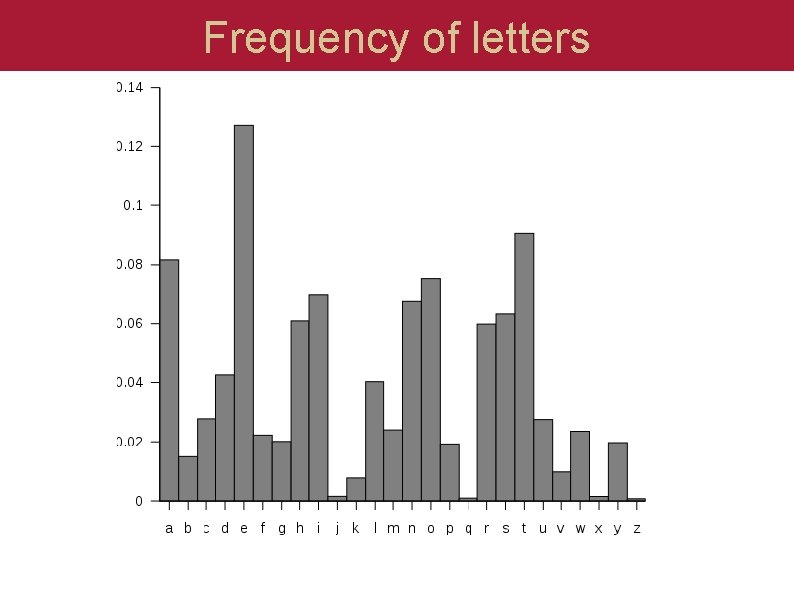
Frequency of letters

Making up sentences If I want to make up a sentence, I could choose some letters at random, based on their occurrence in the alphabet (i. e their scrabble score) rla bsht es stsfa ohhofsd

Lets get clever! What follows a period (“. ”)? Usually a space “ ” What follows a t? Usually an “i” (-tion, -tize, . . . )

Frequency of two letters When the first letter is “t” (from 3, 269 words): ti 51% te 20% ta 15% th 8%

Level 1 analysis Choose a letter based on the probability that it follows the letter before: sha nd t uc t hi ney me l e ol l d

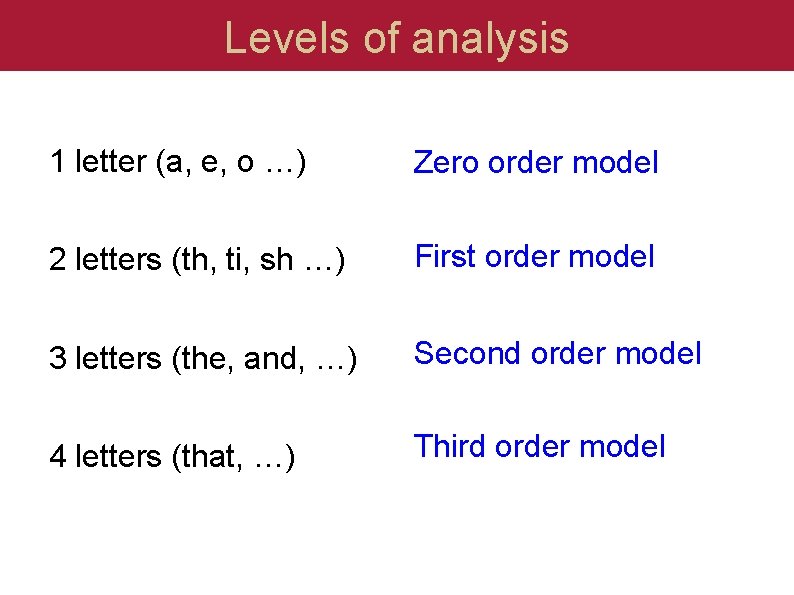
Levels of analysis 1 letter (a, e, o …) Zero order model 2 letters (th, ti, sh …) First order model 3 letters (the, and, …) Second order model 4 letters (that, …) Third order model

Markov models With about 10 th order Markov models of English you get complete words and sentences!

Markov models With about 10 th order Markov models of English you get complete words and sentences!

Markov Models and ORF calling Codons have three letters (ATG, CAC, GGG, . . . ) Use a 2 nd order Markov model for ORF calling The frequency of a letter is predicted based on the frequency of the two letters before

Scrabble

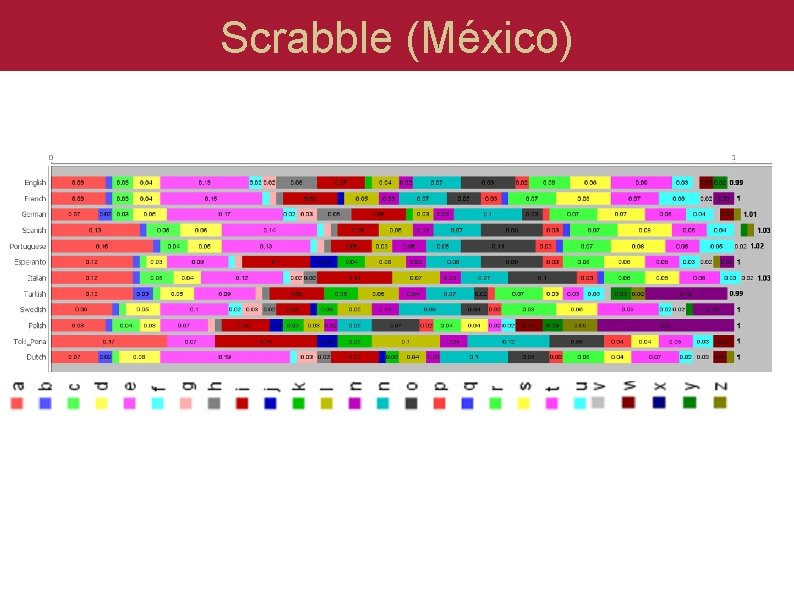
Scrabble (México) Do English and Spanish use the same letters?

Scrabble (México)

Scrabble (US) 1 point: E, A, I, O, N, R, T, L, S, U 2 points: D, G 3 points: B, C, M, P 4 points: F, H, V, W, Y 5 points: K 8 points: J, X 10 points: Q, Z Based on the front page of the NY Times!

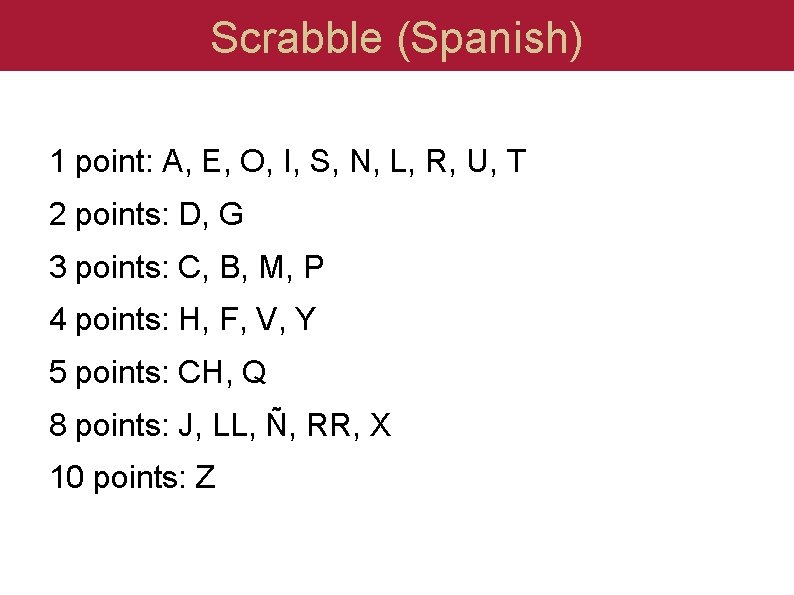
Scrabble (Spanish) 1 point: A, E, O, I, S, N, L, R, U, T 2 points: D, G 3 points: C, B, M, P 4 points: H, F, V, Y 5 points: CH, Q 8 points: J, LL, Ñ, RR, X 10 points: Z

What about scrabble scores for DNA? Will vary with the composition of the organism! Remember, some organisms have high G+C compared to A+T

Markov Models and ORF calling Use a 2 nd order Markov model for ORF calling The frequency of a letter is predicted based on the frequency of the two letters before

Problems! Need to train the Markov model – not all organisms are the same Can use phylogentically close organisms Can use “long orfs” – likely to be correct because unlikely to be random stretches without a stop codon!

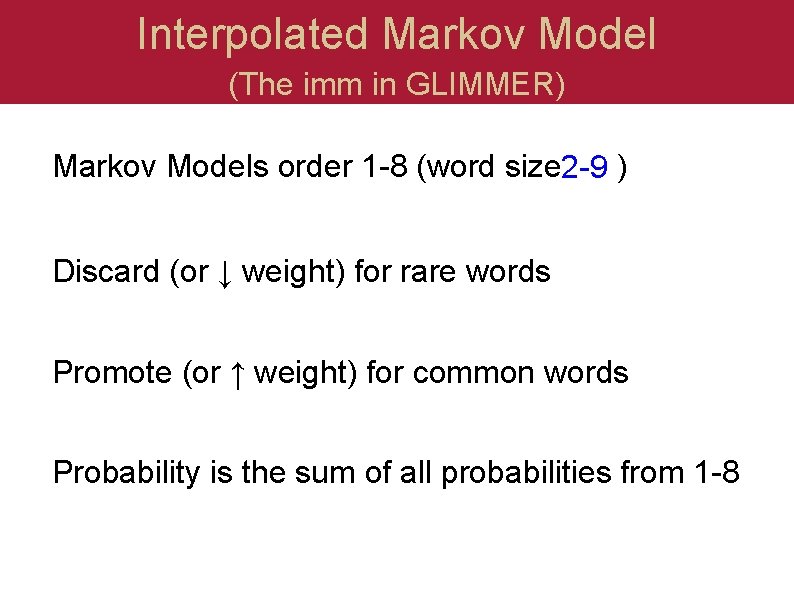
Interpolated Markov Model (The imm in GLIMMER) Markov Models order 1 -8 (word size 2 -9) Discard (or ↓ weight) for rare words Promote (or ↑ weight) for common words Probability is the sum of all probabilities from 1 -8

RNA genes As with proteins, two main methods: Ab initio • Intrinsic Homology based • extrinsic

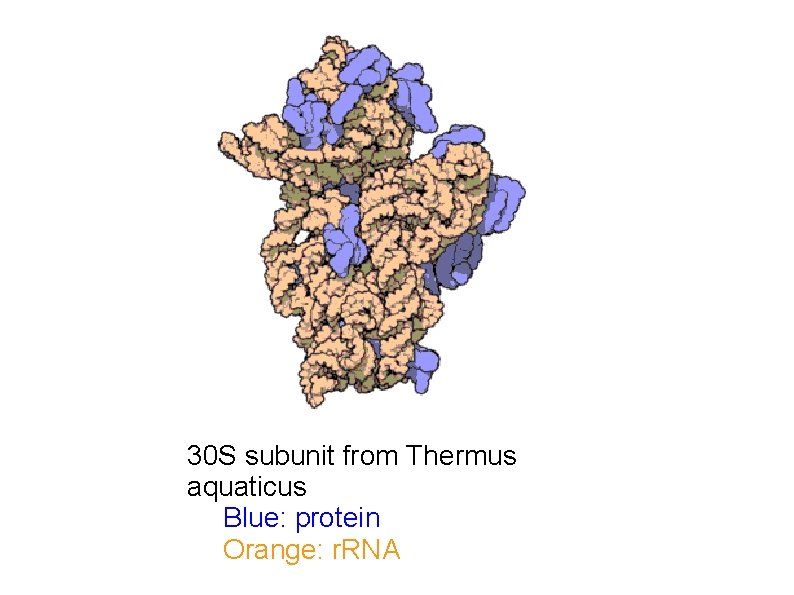
Ribosomes are made of proteins and RNA

30 S subunit from Thermus aquaticus Blue: protein Orange: r. RNA

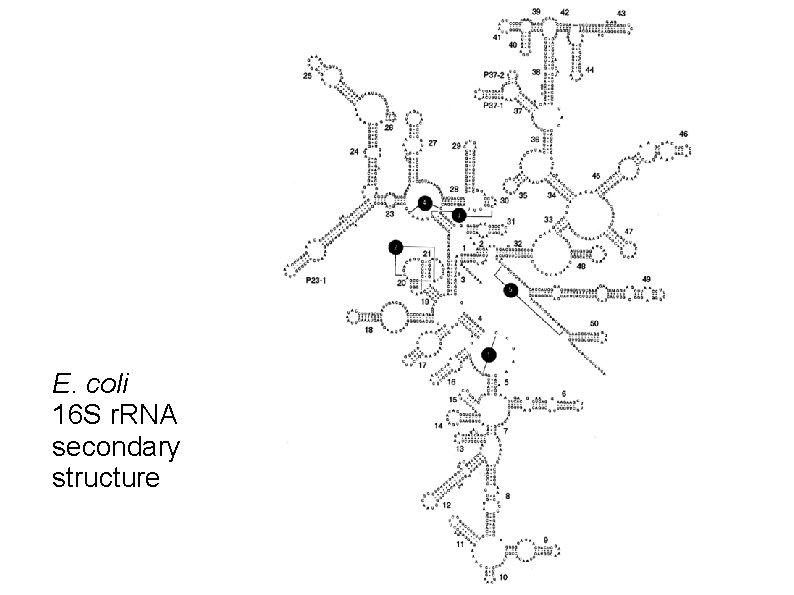
E. coli 16 S r. RNA secondary structure

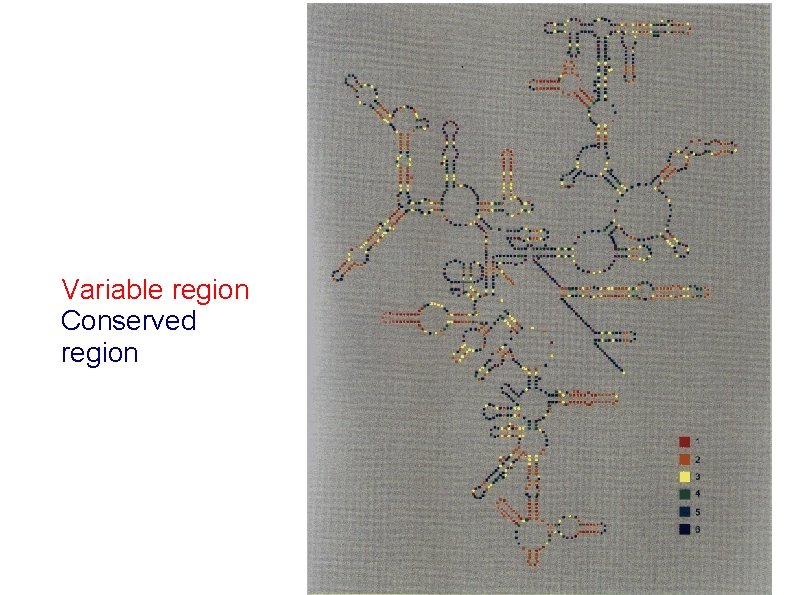
Variable region Conserved region

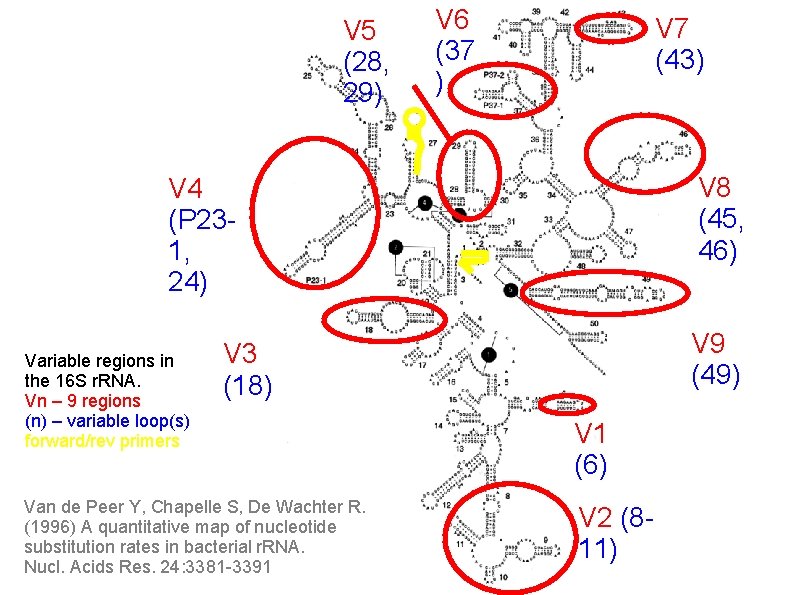
V 5 (28, 29) V 6 (37 ) V 7 (43) V 8 (45, 46) V 4 (P 231, 24) Variable regions in the 16 S r. RNA. Vn – 9 regions (n) – variable loop(s) forward/rev primers V 9 (49) V 3 (18) Van de Peer Y, Chapelle S, De Wachter R. (1996) A quantitative map of nucleotide substitution rates in bacterial r. RNA. Nucl. Acids Res. 24: 3381 -3391 V 1 (6) V 2 (811)

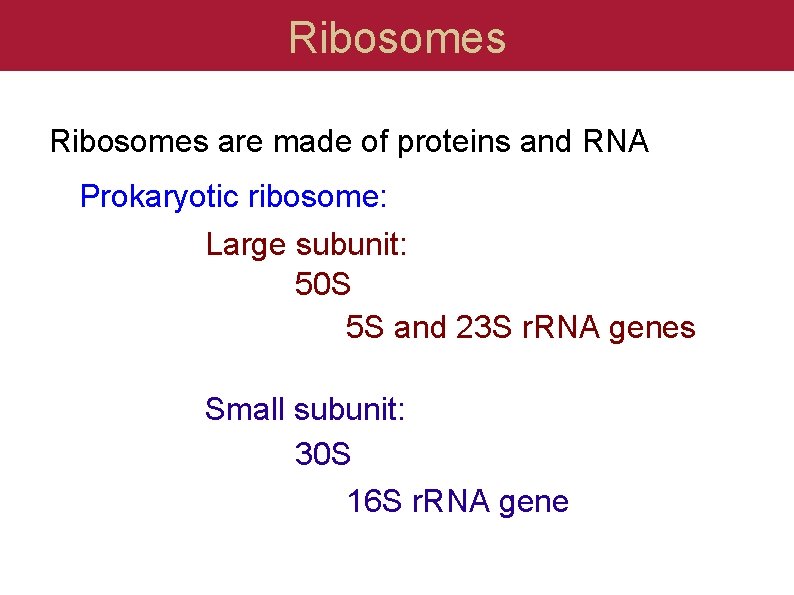
Ribosomes are made of proteins and RNA Prokaryotic ribosome: Large subunit: 50 S 5 S and 23 S r. RNA genes Small subunit: 30 S 16 S r. RNA gene

Finding 16 S genes Easiest way is iterative: BLAST ALIGN TRIM Problem: secondary structure makes identification of the ends difficult

Finding t. RNA genes Not as easy as r. RNA Much shorter Varied sequence Only conservation is 2° structure

t. RNAScan-SE Sean Eddy Use it!

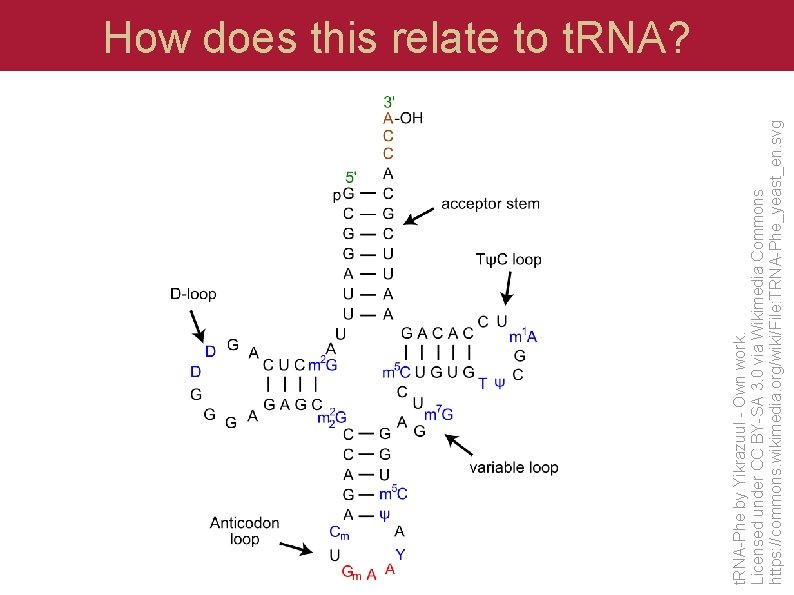
t. RNA-Phe by Yikrazuul - Own work. Licensed under CC BY-SA 3. 0 via Wikimedia Commons https: //commons. wikimedia. org/wiki/File: TRNA-Phe_yeast_en. svg How does this relate to t. RNA?

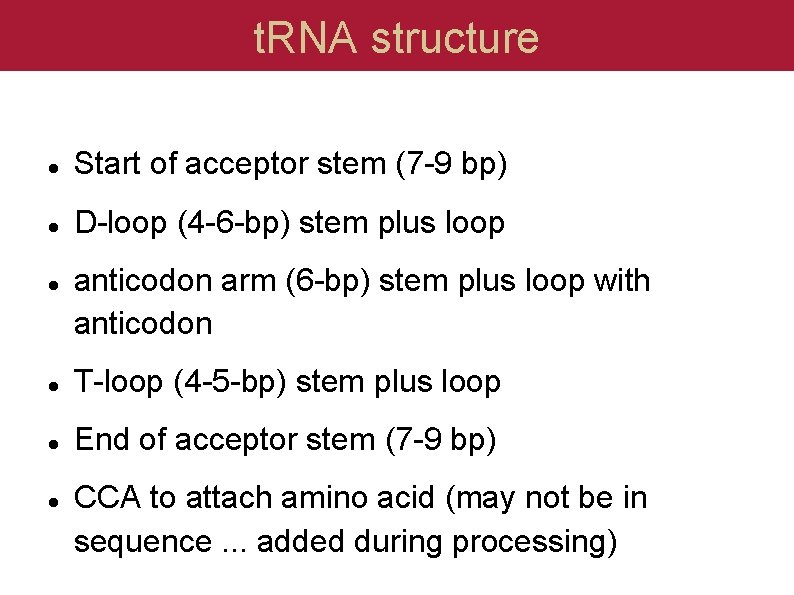
t. RNA structure Start of acceptor stem (7 -9 bp) D-loop (4 -6 -bp) stem plus loop anticodon arm (6 -bp) stem plus loop with anticodon T-loop (4 -5 -bp) stem plus loop End of acceptor stem (7 -9 bp) CCA to attach amino acid (may not be in sequence. . . added during processing)