Orbitrap mass spectrometry a highconfidence screening tool in
Orbitrap mass spectrometry, a highconfidence screening tool in biopharmaceutical product development David Horn Thermo Fisher Scientific San Jose, CA
Outline 1. Challenges in the characterization of biopharmaceuticals 2. Introduction to the new generation of Orbitrap. TM mass spectrometers 3. Confident intact antibody analysis using the Q Exactive. TM Hybrid Mass Spectrometer 4. Introducing Protein Deconvolution 1. 0 software 5. Confident antibody analysis using the Orbitrap Elite. TM Hybrid Mass Spectrometer 6. Summary and conclusions 2
Outline 1. Challenges in the characterization of biopharmaceuticals 2. Introduction to the new generation of Orbitrap. TM mass spectrometers 3. Confident intact antibody analysis using the Q Exactive. TM Hybrid Mass Spectrometer 4. Introducing Protein Deconvolution 1. 0 software 5. High resolution analysis of antibody subunits using the Orbitrap Elite. TM Hybrid Mass Spectrometer 6. Summary and conclusions 3
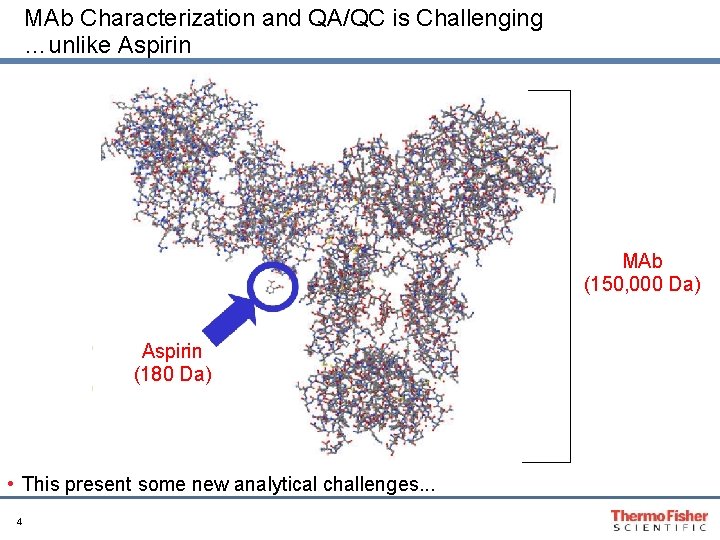
MAb Characterization and QA/QC is Challenging …unlike Aspirin MAb (150, 000 Da) Aspirin (180 Da) • This present some new analytical challenges. . . 4
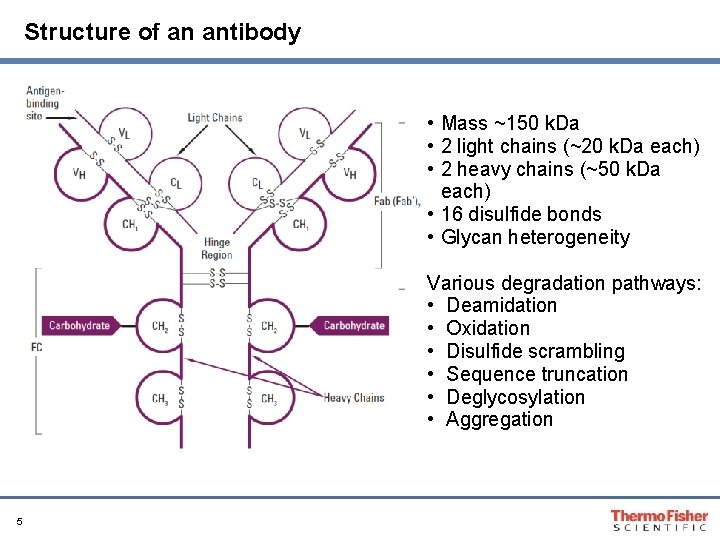
Structure of an antibody • Mass ~150 k. Da • 2 light chains (~20 k. Da each) • 2 heavy chains (~50 k. Da each) • 16 disulfide bonds • Glycan heterogeneity Various degradation pathways: • Deamidation • Oxidation • Disulfide scrambling • Sequence truncation • Deglycosylation • Aggregation 5
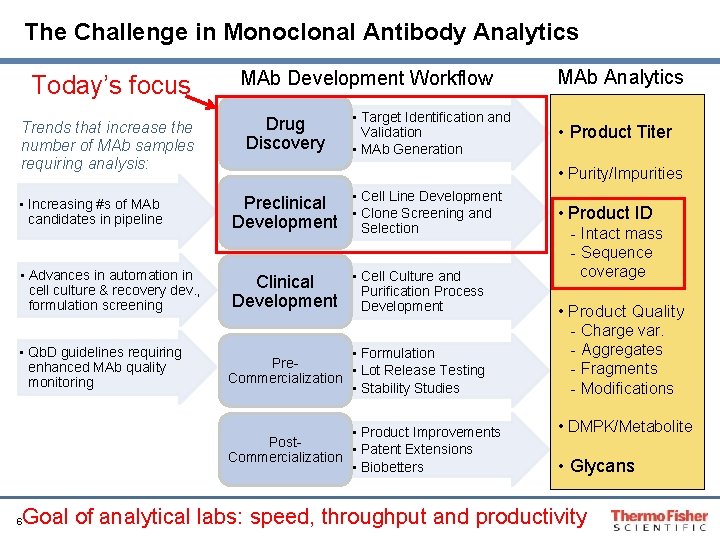
The Challenge in Monoclonal Antibody Analytics Today’s focus Trends that increase the number of MAb samples requiring analysis: MAb Development Workflow Drug Discovery • Target Identification and Validation • MAb Generation Preclinical • Cell Line Development • Clone Screening and Development Selection • Advances in automation in cell culture & recovery dev. , formulation screening • Cell Culture and Clinical Purification Process Development • Formulation Pre • Lot Release Testing Commercialization • Stability Studies • Product Improvements Post • Patent Extensions Commercialization • Biobetters 6 • Product Titer • Purity/Impurities • Increasing #s of MAb candidates in pipeline • Qb. D guidelines requiring enhanced MAb quality monitoring MAb Analytics • Product ID - Intact mass - Sequence coverage • Product Quality - Charge var. - Aggregates - Fragments - Modifications • DMPK/Metabolite • Glycans Goal of analytical labs: speed, throughput and productivity
Thermo Scientific + Dionex: Combining Best-in-Class Technologies Best-in class consumables for protein science Unique ion-chromatography solutions Leading HR/AM mass spectrometers High resolution protein separation columns Bio-HPLC & UHPLC+ Platforms for Protein & MAb Characterization & QA/QC Leading chromatography & MS data systems Today’s Focus 7
Outline 1. Challenges in the characterization of biopharmaceuticals 2. Introduction to the new generation of Orbitrap. TM mass spectrometers 3. Confident intact antibody analysis using the Q Exactive. TM Hybrid Mass Spectrometer 4. Introducing Protein Deconvolution 1. 0 software 5. Confident antibody analysis using the Orbitrap Elite. TM Hybrid Mass Spectrometer 6. Summary and conclusions 8

A new season in life of Orbitrap mass spectrometry 9
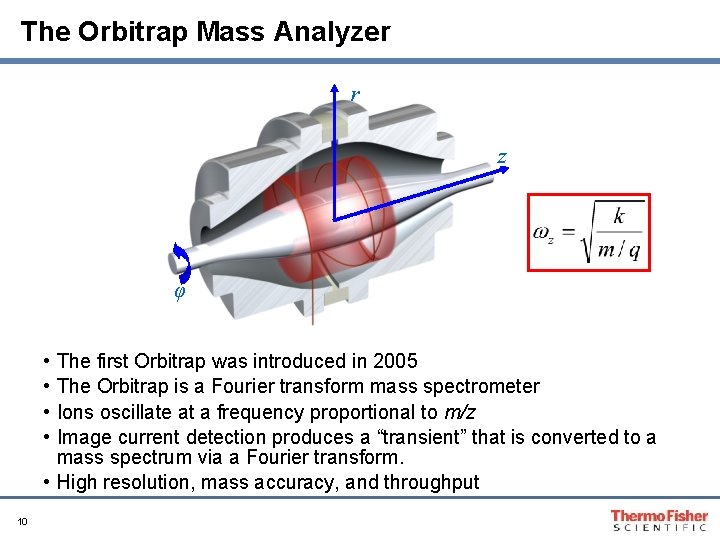
The Orbitrap Mass Analyzer r z φ • • The first Orbitrap was introduced in 2005 The Orbitrap is a Fourier transform mass spectrometer Ions oscillate at a frequency proportional to m/z Image current detection produces a “transient” that is converted to a mass spectrum via a Fourier transform. • High resolution, mass accuracy, and throughput 10
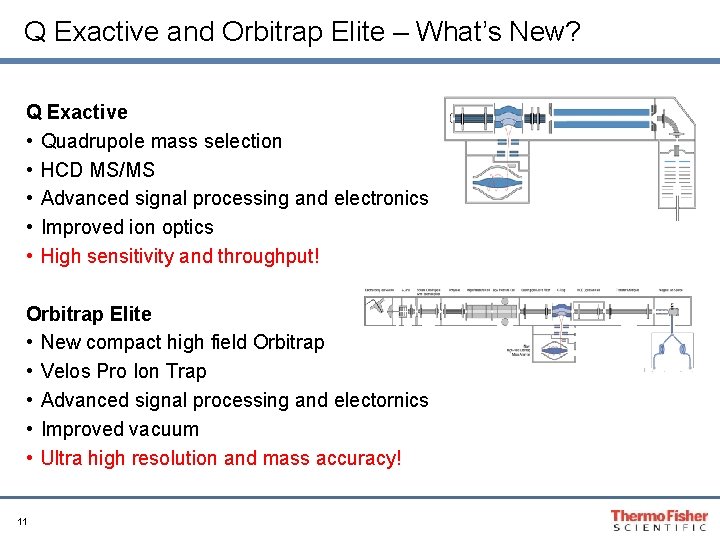
Q Exactive and Orbitrap Elite – What’s New? Q Exactive • Quadrupole mass selection • HCD MS/MS • Advanced signal processing and electronics • Improved ion optics • High sensitivity and throughput! Orbitrap Elite • New compact high field Orbitrap • Velos Pro Ion Trap • Advanced signal processing and electornics • Improved vacuum • Ultra high resolution and mass accuracy! 11
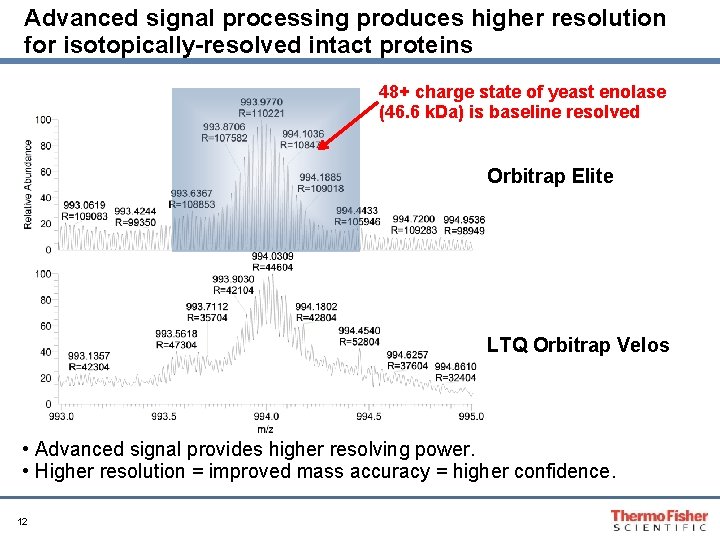
Advanced signal processing produces higher resolution for isotopically-resolved intact proteins 48+ charge state of yeast enolase (46. 6 k. Da) is baseline resolved Orbitrap Elite LTQ Orbitrap Velos • Advanced signal provides higher resolving power. • Higher resolution = improved mass accuracy = higher confidence. 12
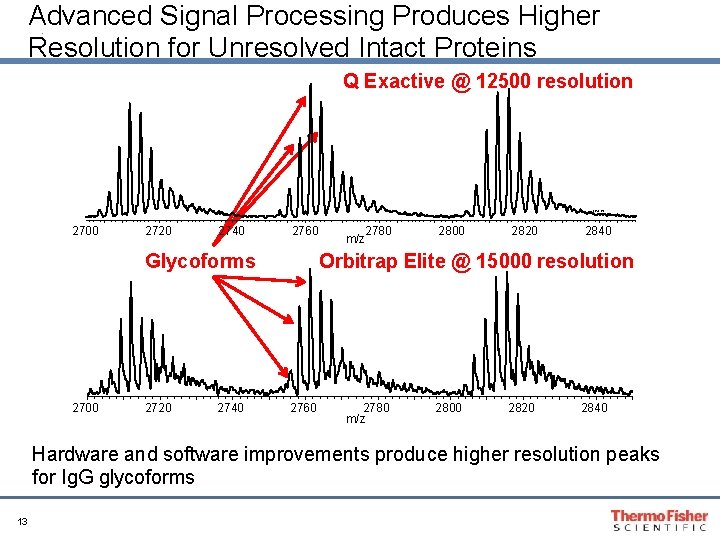
Advanced Signal Processing Produces Higher Resolution for Unresolved Intact Proteins Q Exactive @ 12500 resolution 2842. 63 2700 2720 2740 2760 Glycoforms 2700 2720 2740 2780 m/z 2800 2820 2840 Orbitrap Elite @ 15000 resolution 2760 2780 m/z 2800 2820 2840 Hardware and software improvements produce higher resolution peaks for Ig. G glycoforms 13

Outline 1. Challenges in the characterization of biopharmaceuticals 2. Introduction to the new generation of Orbitrap. TM mass spectrometers 3. Confident intact antibody analysis using the Q Exactive. TM Hybrid Mass Spectrometer 4. Introducing Protein Deconvolution 1. 0 software 5. High resolution analysis of antibody subunits using the Orbitrap Elite. TM Hybrid Mass Spectrometer 6. Summary and conclusions 14
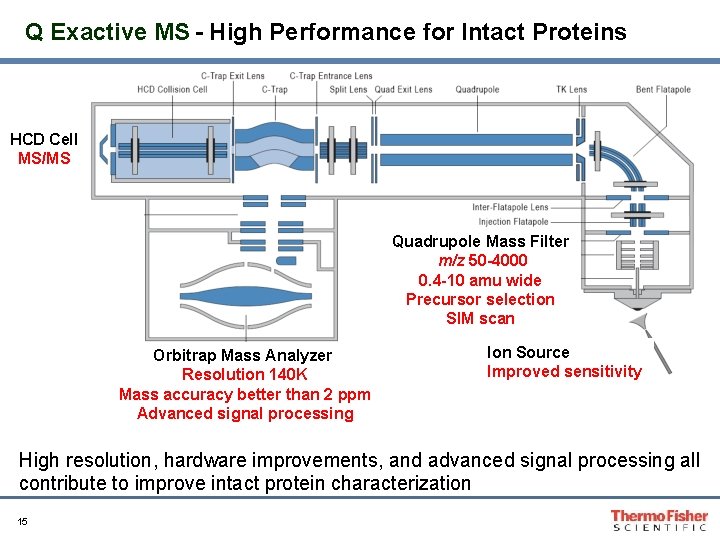
Q Exactive MS - High Performance for Intact Proteins HCD Cell MS/MS Quadrupole Mass Filter m/z 50 -4000 0. 4 -10 amu wide Precursor selection SIM scan Orbitrap Mass Analyzer Resolution 140 K Mass accuracy better than 2 ppm Advanced signal processing Ion Source Improved sensitivity High resolution, hardware improvements, and advanced signal processing all contribute to improve intact protein characterization 15
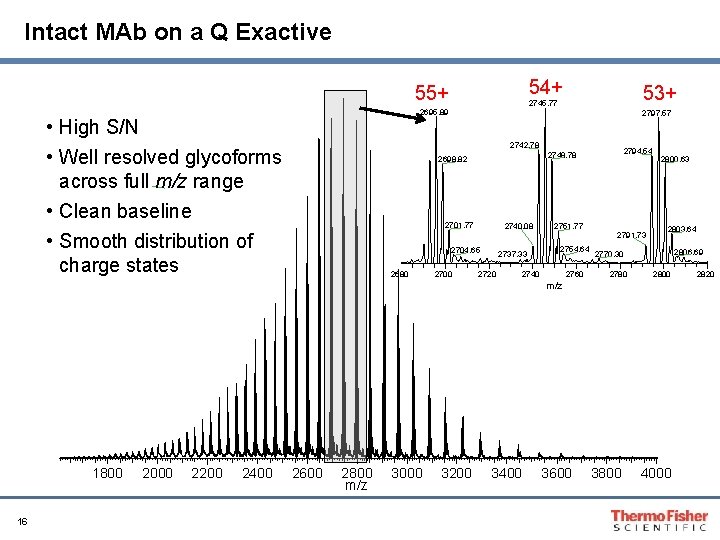
Intact MAb on a Q Exactive 54+ 55+ 2695. 89 • High S/N • Well resolved glycoforms across full m/z range • Clean baseline • Smooth distribution of charge states 53+ 2745. 77 2797. 57 2742. 78 2701. 77 2740. 08 2704. 65 2680 2700 2794. 54 2748. 78 2698. 82 2737. 33 2720 2751. 77 2754. 64 2740 2760 2791. 73 2800. 63 2803. 64 2806. 69 2770. 30 2780 2800 m/z 1800 16 2000 2200 2400 2600 2800 m/z 3000 3200 3400 3600 3800 4000 2820
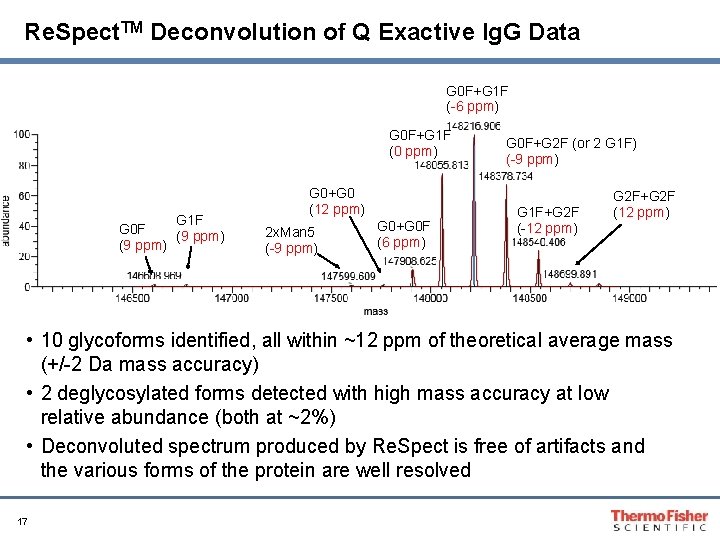
Re. Spect. TM Deconvolution of Q Exactive Ig. G Data G 0 F+G 1 F (-6 ppm) G 0 F+G 1 F (0 ppm) G 0 F (9 ppm) G 1 F (9 ppm) G 0+G 0 (12 ppm) 2 x. Man 5 (-9 ppm) G 0+G 0 F (6 ppm) G 0 F+G 2 F (or 2 G 1 F) (-9 ppm) G 1 F+G 2 F (-12 ppm) G 2 F+G 2 F (12 ppm) • 10 glycoforms identified, all within ~12 ppm of theoretical average mass (+/-2 Da mass accuracy) • 2 deglycosylated forms detected with high mass accuracy at low relative abundance (both at ~2%) • Deconvoluted spectrum produced by Re. Spect is free of artifacts and the various forms of the protein are well resolved 17
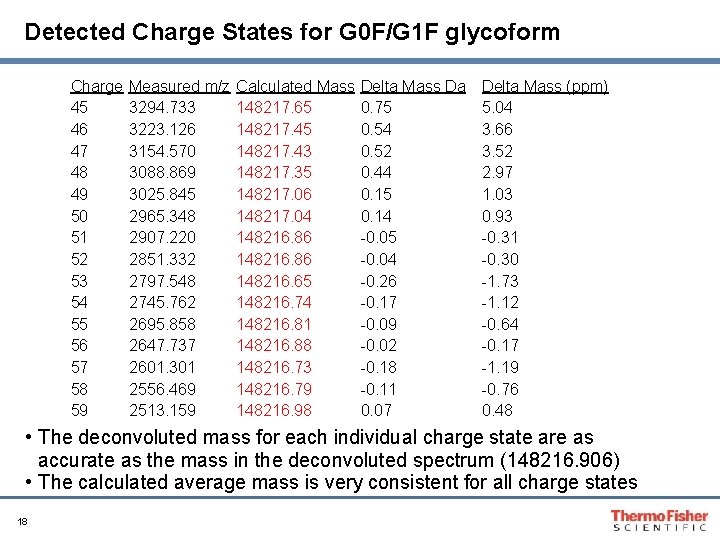
Detected Charge States for G 0 F/G 1 F glycoform Charge 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 Measured m/z 3294. 733 3223. 126 3154. 570 3088. 869 3025. 845 2965. 348 2907. 220 2851. 332 2797. 548 2745. 762 2695. 858 2647. 737 2601. 301 2556. 469 2513. 159 Calculated Mass 148217. 65 148217. 43 148217. 35 148217. 06 148217. 04 148216. 86 148216. 65 148216. 74 148216. 81 148216. 88 148216. 73 148216. 79 148216. 98 Delta Mass Da 0. 75 0. 54 0. 52 0. 44 0. 15 0. 14 -0. 05 -0. 04 -0. 26 -0. 17 -0. 09 -0. 02 -0. 18 -0. 11 0. 07 Delta Mass (ppm) 5. 04 3. 66 3. 52 2. 97 1. 03 0. 93 -0. 31 -0. 30 -1. 73 -1. 12 -0. 64 -0. 17 -1. 19 -0. 76 0. 48 • The deconvoluted mass for each individual charge state are as accurate as the mass in the deconvoluted spectrum (148216. 906) • The calculated average mass is very consistent for all charge states 18
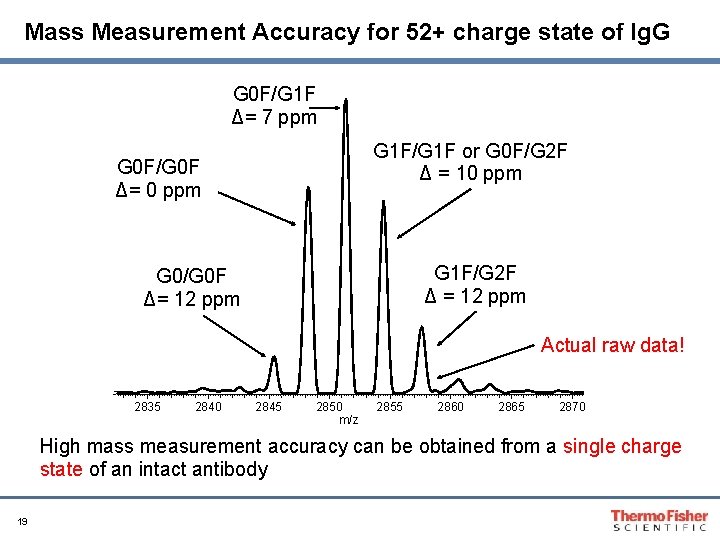
Mass Measurement Accuracy for 52+ charge state of Ig. G G 0 F/G 1 F Δ= 7 ppm G 1 F/G 1 F or G 0 F/G 2 F Δ = 10 ppm G 0 F/G 0 F Δ= 0 ppm G 1 F/G 2 F Δ = 12 ppm G 0/G 0 F Δ= 12 ppm Actual raw data! 2835 2840 2845 2850 m/z 2855 2860 2865 2870 High mass measurement accuracy can be obtained from a single charge state of an intact antibody 19
Mass and Abundance Reproducibility of Ig. G data on a Q Exactive • For the same Ig. G sample as previously shown, 7 different LC/MS runs with various 10 minute LC gradients • Two different Q Exactive instruments were used • Data were acquired on 3 different days • Deconvoluted mass spectra were produced using Protein Deconvolution 1. 0 software • Reproducibility in mass and relative abundance were determined for the 5 major glycoforms of Ig. G 20
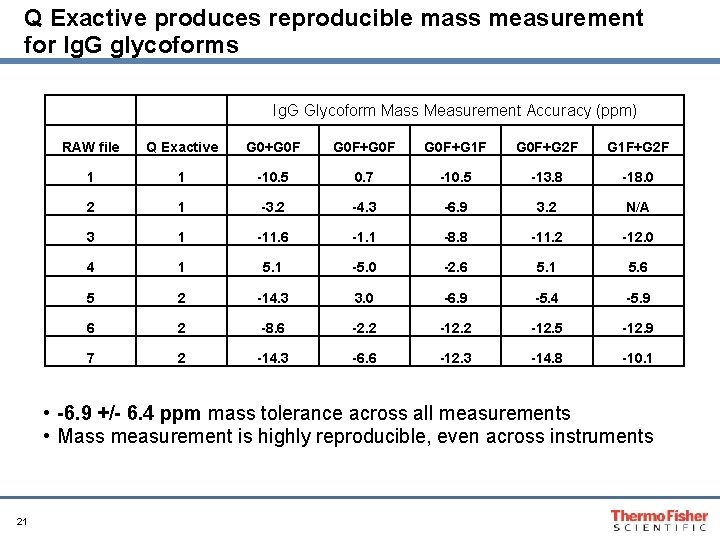
Q Exactive produces reproducible mass measurement for Ig. G glycoforms Ig. G Glycoform Mass Measurement Accuracy (ppm) RAW file Q Exactive G 0+G 0 F+G 1 F G 0 F+G 2 F G 1 F+G 2 F 1 1 -10. 5 0. 7 -10. 5 -13. 8 -18. 0 2 1 -3. 2 -4. 3 -6. 9 3. 2 N/A 3 1 -11. 6 -1. 1 -8. 8 -11. 2 -12. 0 4 1 5. 1 -5. 0 -2. 6 5. 1 5. 6 5 2 -14. 3 3. 0 -6. 9 -5. 4 -5. 9 6 2 -8. 6 -2. 2 -12. 5 -12. 9 7 2 -14. 3 -6. 6 -12. 3 -14. 8 -10. 1 • -6. 9 +/- 6. 4 ppm mass tolerance across all measurements • Mass measurement is highly reproducible, even across instruments 21
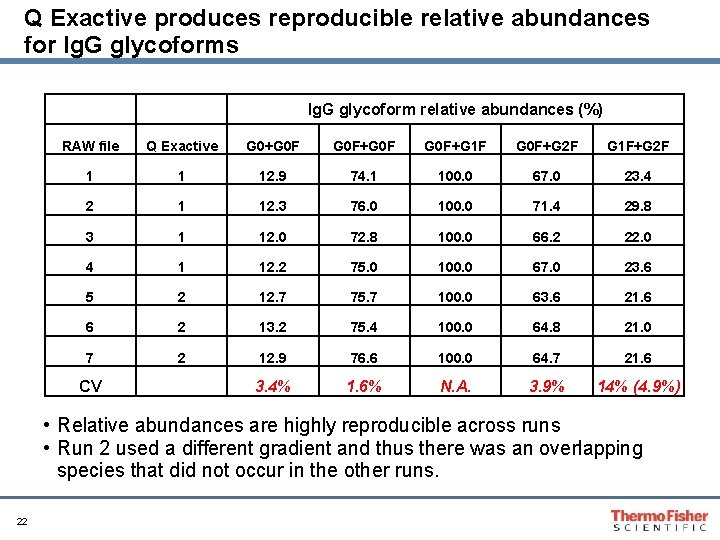
Q Exactive produces reproducible relative abundances for Ig. G glycoforms Ig. G glycoform relative abundances (%) RAW file Q Exactive G 0+G 0 F+G 1 F G 0 F+G 2 F G 1 F+G 2 F 1 1 12. 9 74. 1 100. 0 67. 0 23. 4 2 1 12. 3 76. 0 100. 0 71. 4 29. 8 3 1 12. 0 72. 8 100. 0 66. 2 22. 0 4 1 12. 2 75. 0 100. 0 67. 0 23. 6 5 2 12. 7 75. 7 100. 0 63. 6 21. 6 6 2 13. 2 75. 4 100. 0 64. 8 21. 0 7 2 12. 9 76. 6 100. 0 64. 7 21. 6 3. 4% 1. 6% N. A. 3. 9% 14% (4. 9%) CV • Relative abundances are highly reproducible across runs • Run 2 used a different gradient and thus there was an overlapping species that did not occur in the other runs. 22
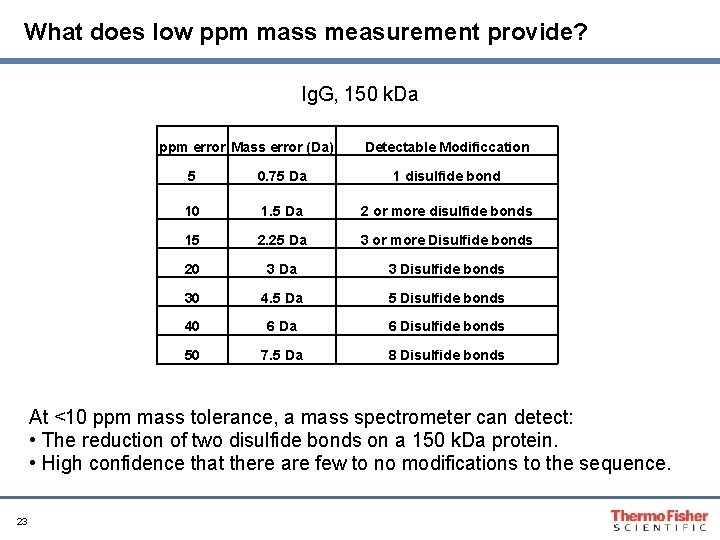
What does low ppm mass measurement provide? Ig. G, 150 k. Da ppm error Mass error (Da) Detectable Modificcation 5 0. 75 Da 1 disulfide bond 10 1. 5 Da 2 or more disulfide bonds 15 2. 25 Da 3 or more Disulfide bonds 20 3 Da 3 Disulfide bonds 30 4. 5 Da 5 Disulfide bonds 40 6 Da 6 Disulfide bonds 50 7. 5 Da 8 Disulfide bonds At <10 ppm mass tolerance, a mass spectrometer can detect: • The reduction of two disulfide bonds on a 150 k. Da protein. • High confidence that there are few to no modifications to the sequence. 23
Q Exactive Summary • The Q Exactive produces very high mass accuracy for intact antibodies • These masses and abundances can be very reproducibly measured • This indicates that the Q Exactive will be excellent for high throughput confirmation of biopharmaceuticals 24
Outline 1. Challenges in the characterization of biopharmaceuticals 2. Introduction to the new generation of Orbitrap. TM mass spectrometers 3. Confident intact antibody analysis using the Q Exactive. TM Hybrid Mass Spectrometer 4. Introducing Protein Deconvolution 1. 0 software 5. High resolution analysis of antibody subunits using the Orbitrap Elite. TM Hybrid Mass Spectrometer 6. Summary and conclusions 25
Protein Deconvolution 1. 0 • Workflow software for intact protein mass determination • Supports all Orbitrap mass spectrometers • Includes 2 deconvolution algorithms: • Xtract for isotopically resolved proteins • Re. Spect for isotopically unresolved proteins (e. g. Ig. G) • Target release date: Early November • For more information – create an account at the Thermo Proteomics Software Portal (http: //portal. thermo-brims. com) Re. Spect is a trademark of Positive Probability, Ltd. 26
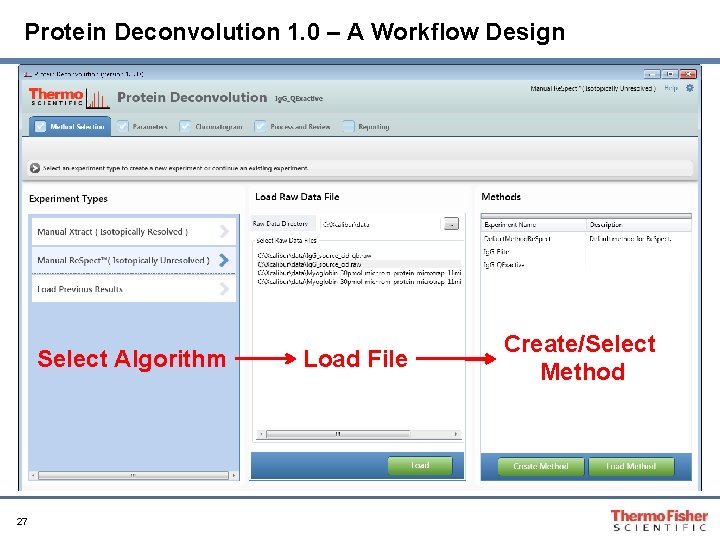
Protein Deconvolution 1. 0 – A Workflow Design Select Algorithm 27 Load File Create/Select Method
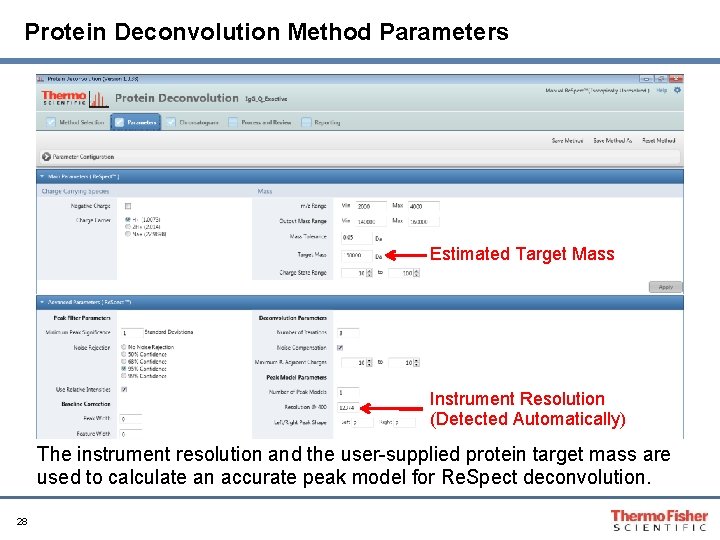
Protein Deconvolution Method Parameters Estimated Target Mass Instrument Resolution (Detected Automatically) The instrument resolution and the user-supplied protein target mass are used to calculate an accurate peak model for Re. Spect deconvolution. 28
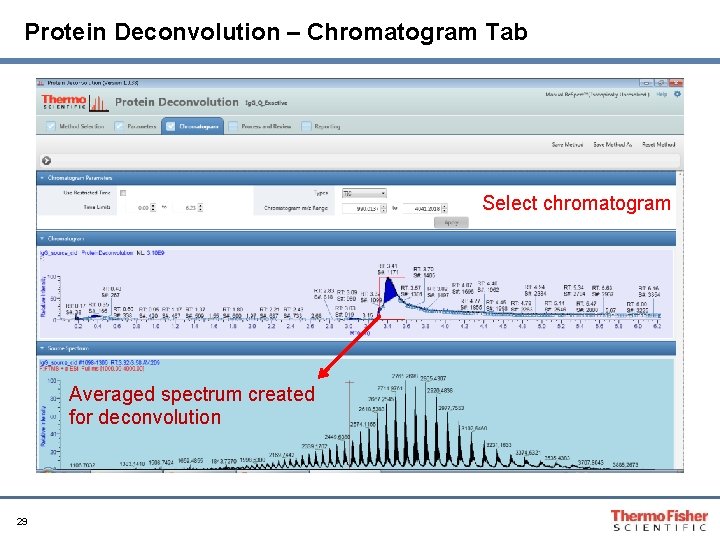
Protein Deconvolution – Chromatogram Tab Select chromatogram Averaged spectrum created for deconvolution 29
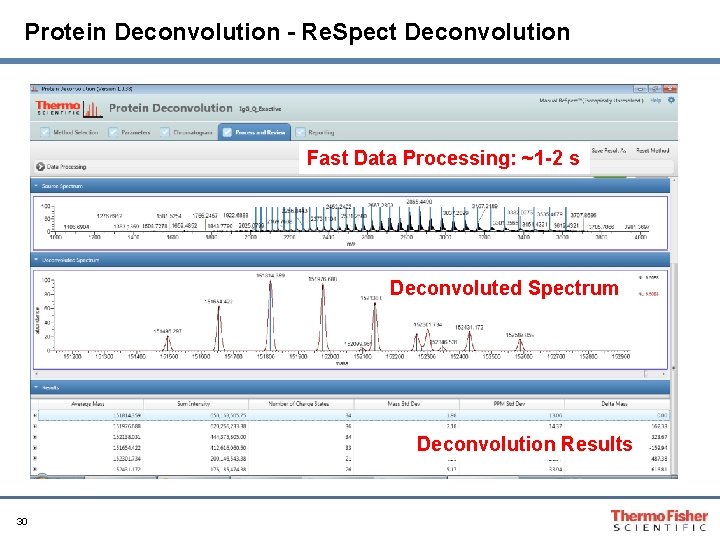
Protein Deconvolution - Re. Spect Deconvolution Fast Data Processing: ~1 -2 s Deconvoluted Spectrum Deconvolution Results 30
Protein Deconvolution - Report Comprehensive, Exportable Report 31
Outline 1. Challenges in the characterization of biopharmaceuticals 2. Introduction to the new generation in Orbitrap. TM mass spectrometers 3. Confident intact antibody analysis using the Q Exactive. TM Hybrid Mass Spectrometer 4. Introducing Protein Deconvolution 1. 0 software 5. High resolution analysis of antibody subunits using the Orbitrap Elite. TM Hybrid Mass Spectrometer 6. Summary and conclusions 32
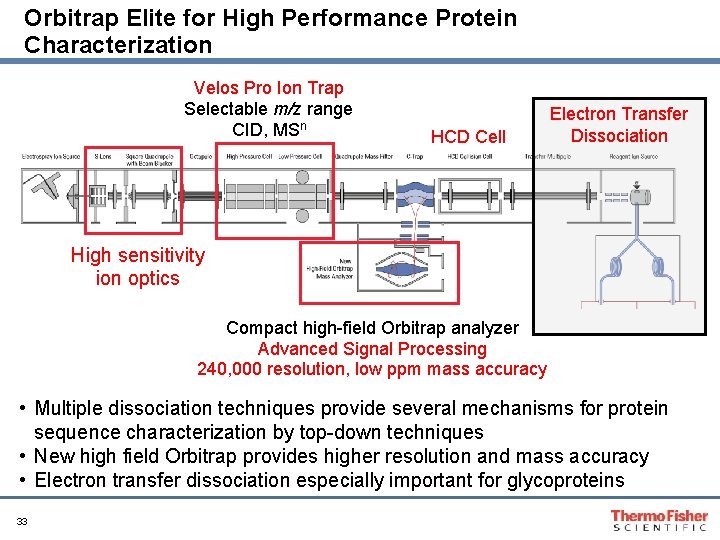
Orbitrap Elite for High Performance Protein Characterization Velos Pro Ion Trap Selectable m/z range CID, MSn HCD Cell Electron Transfer Dissociation High sensitivity ion optics Compact high-field Orbitrap analyzer Advanced Signal Processing 240, 000 resolution, low ppm mass accuracy • Multiple dissociation techniques provide several mechanisms for protein sequence characterization by top-down techniques • New high field Orbitrap provides higher resolution and mass accuracy • Electron transfer dissociation especially important for glycoproteins 33
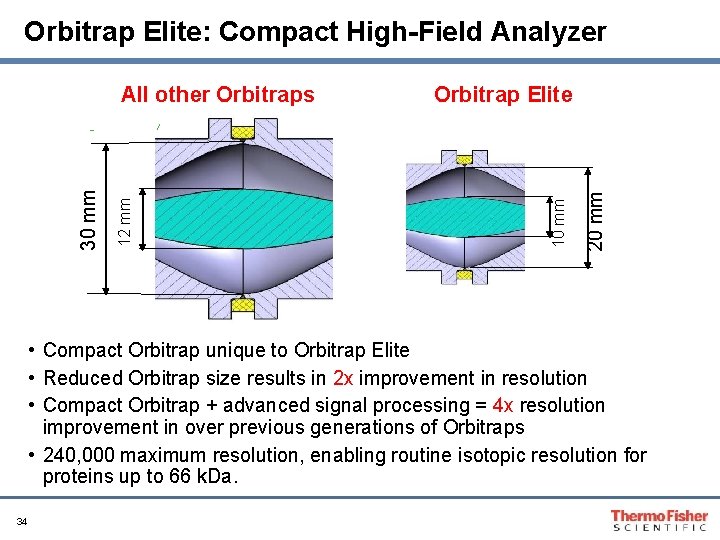
Orbitrap Elite: Compact High-Field Analyzer 20 mm Orbitrap Elite 10 mm 12 mm 30 mm All other Orbitraps • Compact Orbitrap unique to Orbitrap Elite • Reduced Orbitrap size results in 2 x improvement in resolution • Compact Orbitrap + advanced signal processing = 4 x resolution improvement in over previous generations of Orbitraps • 240, 000 maximum resolution, enabling routine isotopic resolution for proteins up to 66 k. Da. 34
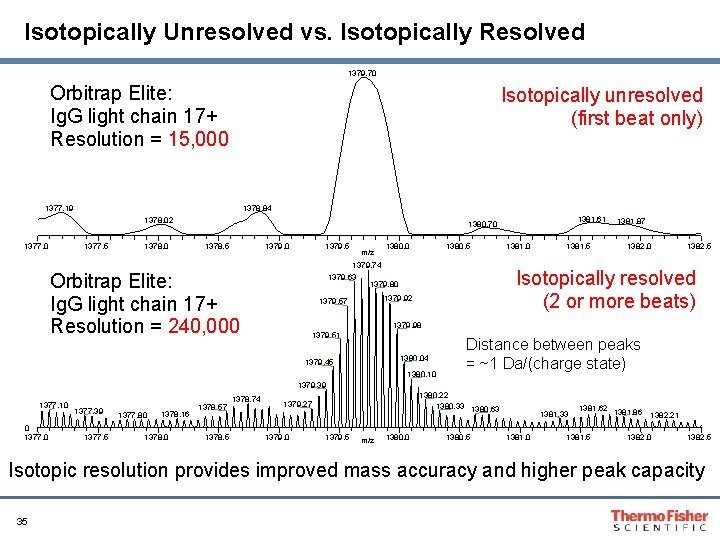
Isotopically Unresolved vs. Isotopically Resolved 1379. 70 Orbitrap Elite: Ig. G light chain 17+ Resolution = 15, 000 Isotopically unresolved (first beat only) 1378. 84 1377. 19 1378. 02 1377. 0 1377. 5 1378. 0 1381. 61 1380. 70 1378. 5 1379. 0 1379. 5 m/z 1380. 0 1380. 5 1379. 74 1379. 63 1379. 80 Orbitrap Elite: Ig. G light chain 17+ Resolution = 240, 000 1381. 5 1382. 0 1382. 5 Isotopically resolved (2 or more beats) 1379. 92 1379. 57 1381. 0 1381. 87 1379. 98 1379. 51 1380. 04 1379. 45 1380. 10 Distance between peaks = ~1 Da/(charge state) 1379. 39 1377. 10 0 1377. 39 1377. 5 1377. 80 1378. 16 1378. 0 1378. 57 1378. 5 1378. 74 1380. 22 1380. 33 1380. 63 1379. 27 1379. 0 1379. 5 m/z 1380. 0 1380. 5 1381. 33 1381. 0 1381. 62 1381. 86 1382. 21 1381. 5 1382. 0 1382. 5 Isotopic resolution provides improved mass accuracy and higher peak capacity 35
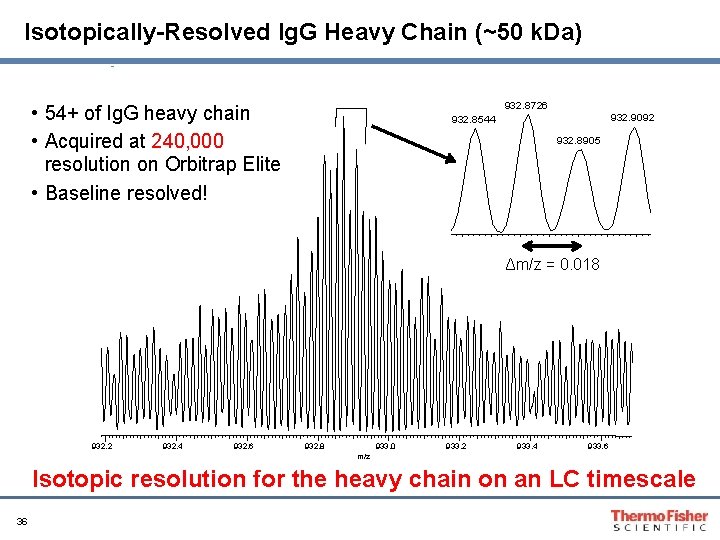
Isotopically-Resolved Ig. G Heavy Chain (~50 k. Da) 932. 8726 • 54+ of Ig. G heavy chain • Acquired at 240, 000 resolution on Orbitrap Elite • Baseline resolved! 932. 9092 932. 8544 932. 8905 Δm/z = 0. 018 932. 2 932. 4 932. 6 932. 8 933. 0 933. 2 933. 4 933. 6 m/z Isotopic resolution for the heavy chain on an LC timescale 36
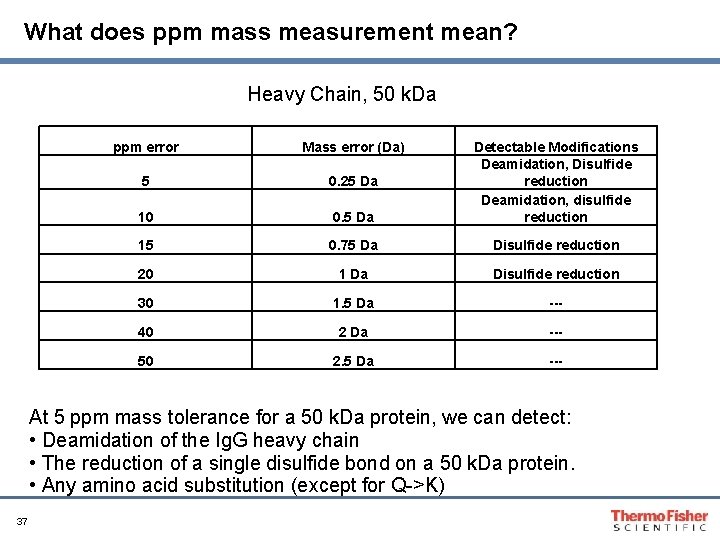
What does ppm mass measurement mean? Heavy Chain, 50 k. Da ppm error Mass error (Da) 5 0. 25 Da 10 0. 5 Da Detectable Modifications Deamidation, Disulfide reduction Deamidation, disulfide reduction 15 0. 75 Da Disulfide reduction 20 1 Da Disulfide reduction 30 1. 5 Da --- 40 2 Da --- 50 2. 5 Da --- At 5 ppm mass tolerance for a 50 k. Da protein, we can detect: • Deamidation of the Ig. G heavy chain • The reduction of a single disulfide bond on a 50 k. Da protein. • Any amino acid substitution (except for Q->K) 37
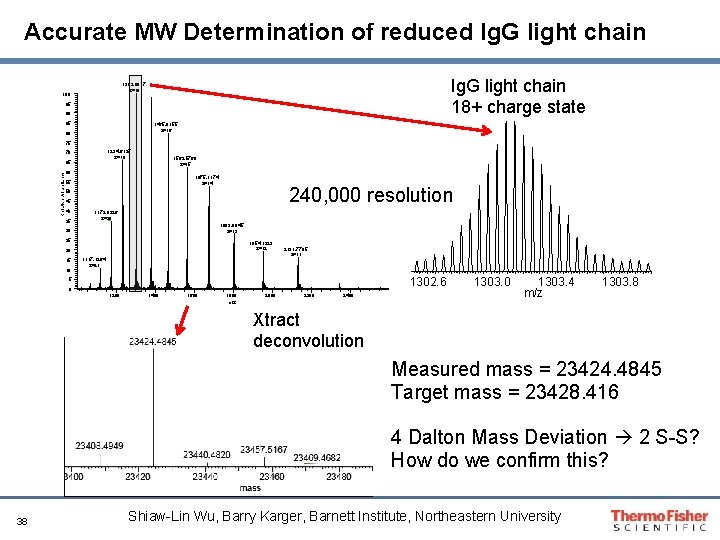
Accurate MW Determination of reduced Ig. G light chain 18+ charge state 1303. 0917 z=18 100 95 90 85 1465. 9155 z=16 80 75 1234. 6137 z=19 70 Relative Abundance 65 1563. 5760 z=15 60 1675. 1174 z=14 55 240, 000 resolution 50 45 40 35 1172. 8326 z=20 1803. 8945 z=13 30 25 1954. 1323 z=12 20 15 10 1117. 1264 z=21 2131. 7785 z=11 1302. 6 5 0 1200 1400 1600 1800 m/z 2000 2200 2400 1303. 4 m/z 1303. 8 Xtract deconvolution Measured mass = 23424. 4845 Target mass = 23428. 416 4 Dalton Mass Deviation 2 S-S? How do we confirm this? 38 Shiaw-Lin Wu, Barry Karger, Barnett Institute, Northeastern University
“Top Down” vs. “Bottom Up” protein analysis • Proteins are usually digested with a proteolytic enzyme and analyzed using peptide mass fingerprinting or data dependent MS/MS • Peptide mass fingerprinting has some disadvantages, including introduction of artifacts into the sample and there is no guarantee of 100% sequence coverage • An alternative strategy is to use a “top down” methodology, where the intact protein is isolated and fragmented in the mass spectrometer using either a data-dependent or targeted acquisition method • High resolution mass spectrometry is a must • References: • Kelleher et al, “Top Down versus Bottom Up Protein Characterization by Tandem High Resolution Mass Spectrometry”, J. Am. Chem. Soc. , 1999, 21, pp. 806 -812 • Bondarenko et al, “Mass Measurement and Top-Down HPLC/MS Analysis of Intact Monoclonal Antibodies on a Hybrid Linear Quadrupole Ion Trap-Orbitrap Mass Spectrometer”, J. Am. Soc. Mass Spectrom. , 2009, 20, pp. 1415 -24. 39
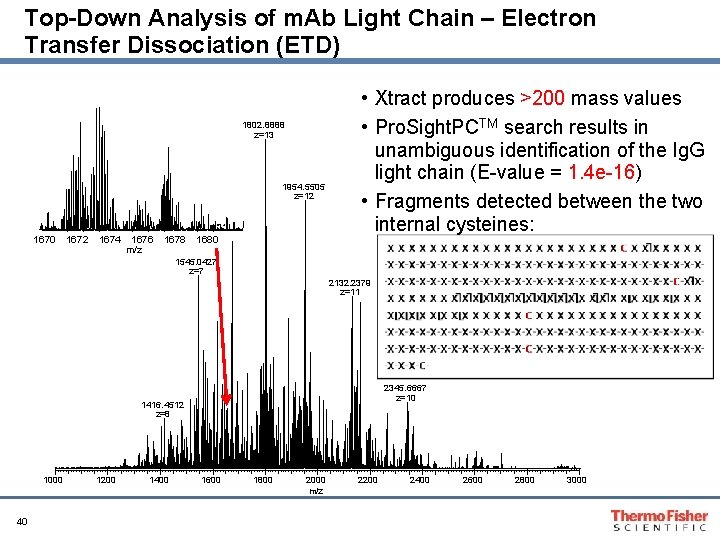
Top-Down Analysis of m. Ab Light Chain – Electron Transfer Dissociation (ETD) 1802. 8888 z=13 1954. 5505 z=12 1680. 45 1670 1672 1674 1676 m/z 1678 1680 • Xtract produces >200 mass values • Pro. Sight. PCTM search results in unambiguous identification of the Ig. G light chain (E-value = 1. 4 e-16) • Fragments detected between the two internal cysteines: 1545. 0427 z=7 2132. 2379 z=11 2345. 6667 z=10 1416. 4512 z=8 1000 40 1200 1400 1600 1800 2000 m/z 2200 2400 2600 2800 3000
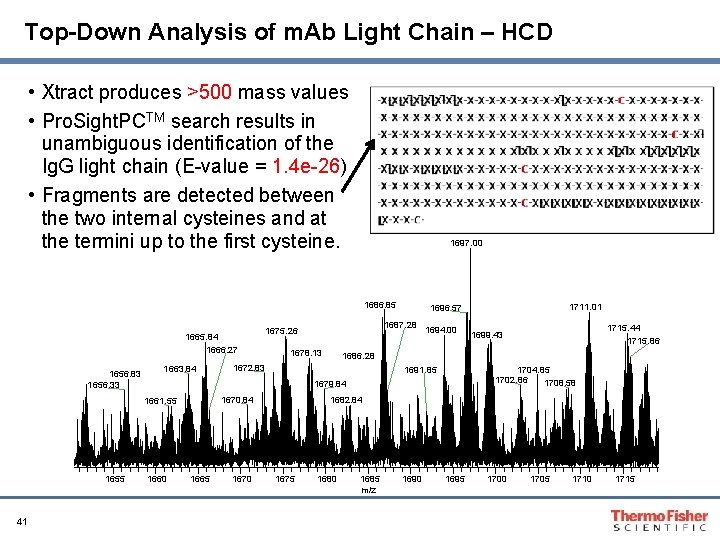
Top-Down Analysis of m. Ab Light Chain – HCD • Xtract produces >500 mass values • Pro. Sight. PCTM search results in unambiguous identification of the Ig. G light chain (E-value = 1. 4 e-26) • Fragments are detected between the two internal cysteines and at the termini up to the first cysteine. 1697. 00 1686. 85 1665. 84 1666. 27 1656. 83 1656. 33 1663. 84 41 1678. 13 1694. 00 1704. 85 1702. 86 1708. 58 1691. 85 1670. 84 1660 1665 1670 1715. 44 1715. 86 1699. 43 1686. 28 1672. 83 1679. 84 1661. 55 1687. 28 1675. 26 1711. 01 1696. 57 1682. 84 1675 1680 1685 m/z 1690 1695 1700 1705 1710 1715
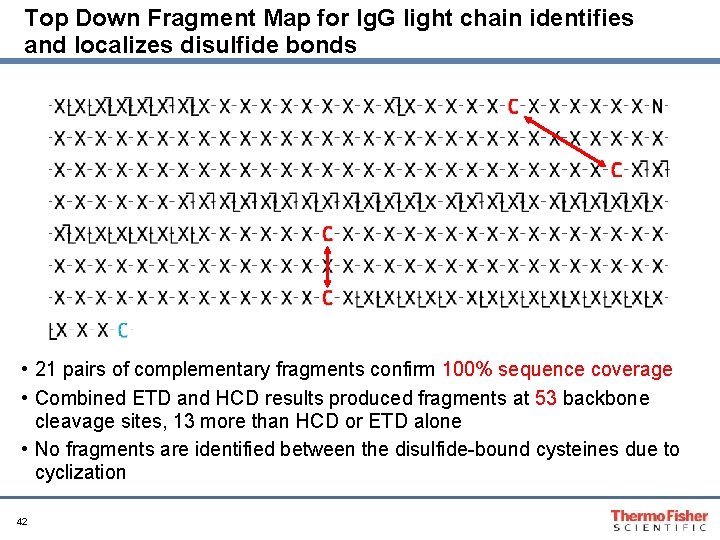
Top Down Fragment Map for Ig. G light chain identifies and localizes disulfide bonds • 21 pairs of complementary fragments confirm 100% sequence coverage • Combined ETD and HCD results produced fragments at 53 backbone cleavage sites, 13 more than HCD or ETD alone • No fragments are identified between the disulfide-bound cysteines due to cyclization 42
Summary – Orbitrap Elite • The Orbitrap Elite is well suited for both intact protein confirmation as well as top down protein characterization • Top down protein characterization is an alternative to bottom-up peptide MS/MS for identification and confirmation of expected and unexpected changes to the target protein 43
Summary and Conclusions • Orbitrap-based systems are excellent for biopharmaceutical characterization • The Q Exactive and Orbitrap Elite are the best Orbitrap systems yet for intact biopharmaceutical analysis • Protein Deconvolution 1. 0 produces highly accurate confident intact protein masses and abundances • Pro. Sight. PC is applicable to biopharmaceutical applications (not just top down proteomics) • High confidence results allow scientists in biopharmaceutical labs to increase sample throughput by bypassing more time-consuming experiments 44
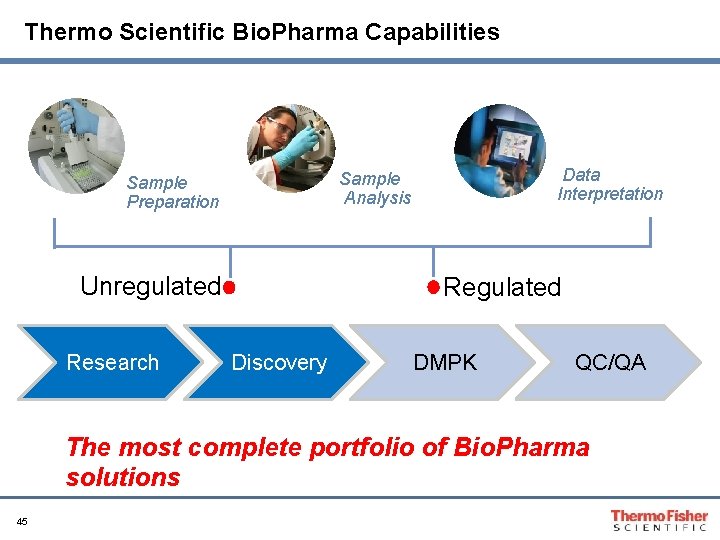
Thermo Scientific Bio. Pharma Capabilities Unregulated Research Data Interpretation Sample Analysis Sample Preparation Regulated Discovery DMPK QC/QA The most complete portfolio of Bio. Pharma solutions 45
Acknowledgements Marketing Andreas Hühmer Thomas Moehring Markus Kellmann Yi Zhang Zhiqi Hao Seema Sharma Rosa Viner Vlad Zabrouskov Amy Zumwalt Shannon Eliuk Reiko Kiyonami Justin Blethrow Julian Saba Scott Peterman 46 Software R&D Steve Chaput Doug Miller Tom Mc. Clure Grace Li Paul Gazis Helen Tran Shijun Li Barbara Gibson Hardware R&D Alexander Makarov Jae Schwartz Martin Zeller

Thank you—Q&A Partners in driving value creation 47
- Slides: 47