Optimizing Stata for Analysis of Large Data Sets










- Slides: 29

Optimizing Stata for Analysis of Large Data Sets Joseph Canner, MHS Eric Schneider, Ph. D Johns Hopkins University Stata Conference New Orleans, LA July 19, 2013

Background • Programmer/Statistician: 20 years experience with SAS • Took new job and started using Stata in January 2013 • Reviewed many do-files from predecessors and colleagues in order to learn Stata and understand new job

Caveats • Large data sets: irrelevant if you don’t use large data sets and/or if you don’t have a system that has sufficient memory to analyze large data sets • Coding practices: these are examples from real users, but not necessarily trained programmers or Stata experts

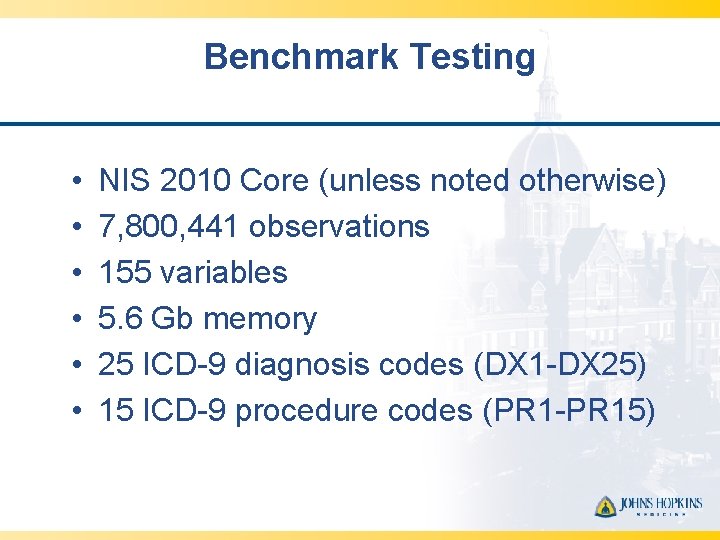
Benchmark Testing • • • NIS 2010 Core (unless noted otherwise) 7, 800, 441 observations 155 variables 5. 6 Gb memory 25 ICD-9 diagnosis codes (DX 1 -DX 25) 15 ICD-9 procedure codes (PR 1 -PR 15)

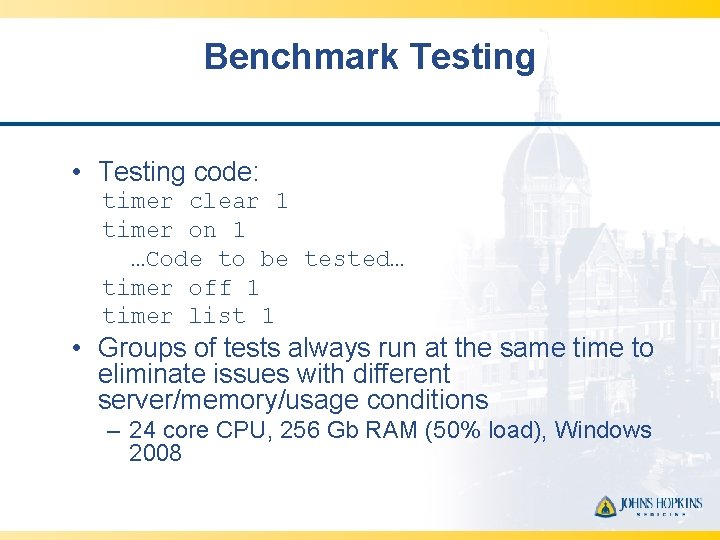
Benchmark Testing • Testing code: timer clear 1 timer on 1 …Code to be tested… timer off 1 timer list 1 • Groups of tests always run at the same time to eliminate issues with different server/memory/usage conditions – 24 core CPU, 256 Gb RAM (50% load), Windows 2008

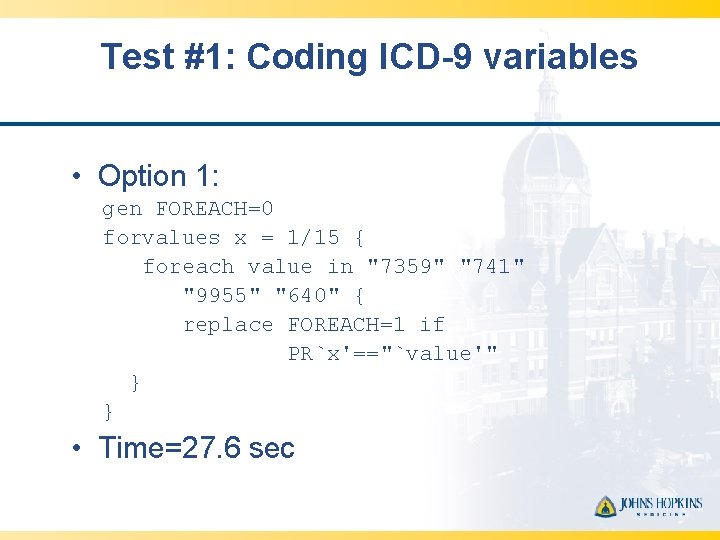
Test #1: Coding ICD-9 variables • Option 1: gen FOREACH=0 forvalues x = 1/15 { foreach value in "7359" "741" "9955" "640" { replace FOREACH=1 if PR`x'=="`value'" } } • Time=27. 6 sec

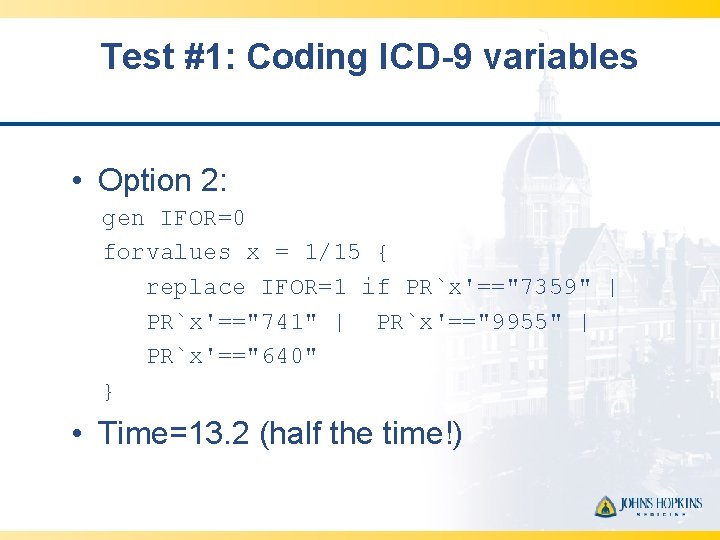
Test #1: Coding ICD-9 variables • Option 2: gen IFOR=0 forvalues x = 1/15 { replace IFOR=1 if PR`x'=="7359" | PR`x'=="741" | PR`x'=="9955" | PR`x'=="640" } • Time=13. 2 (half the time!)

Test #1: Coding ICD-9 variables • Option 3: gen INLIST=0 forvalues x = 1/15 { replace INLIST=1 if inlist(PR`x', "7359", "741", "9955", "640") } • Time=9. 6 sec (a little better than Option 2, and easier to write and read)

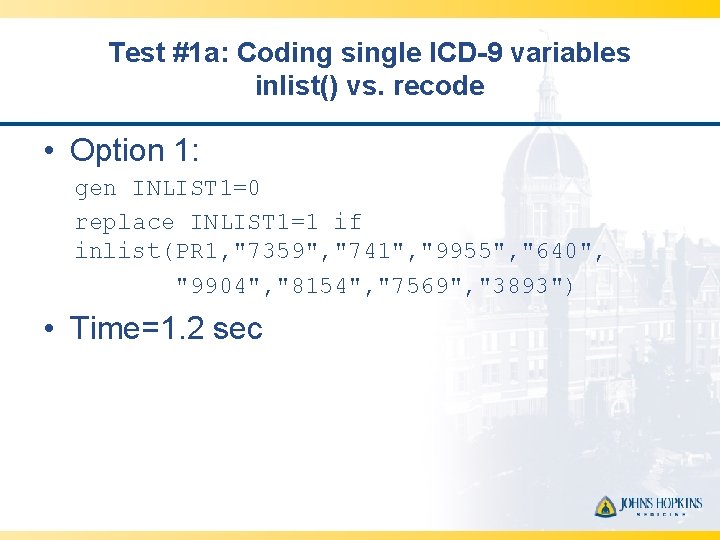
Test #1 a: Coding single ICD-9 variables inlist() vs. recode • Option 1: gen INLIST 1=0 replace INLIST 1=1 if inlist(PR 1, "7359", "741", "9955", "640", "9904", "8154", "7569", "3893") • Time=1. 2 sec

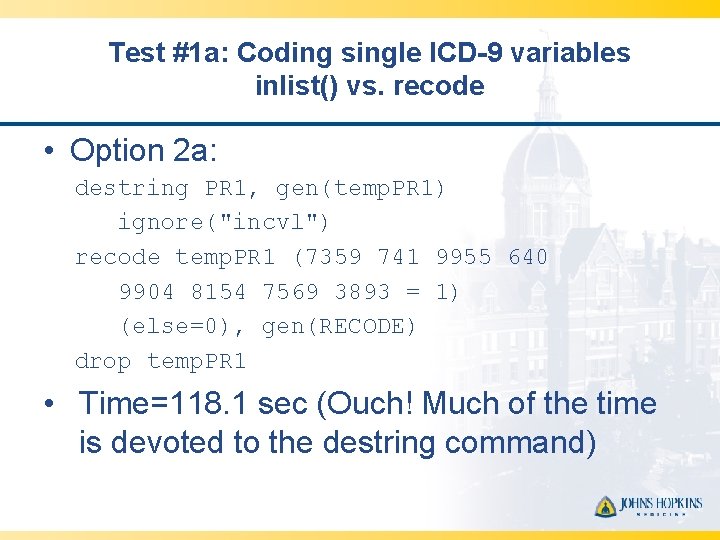
Test #1 a: Coding single ICD-9 variables inlist() vs. recode • Option 2 a: destring PR 1, gen(temp. PR 1) ignore("incvl") recode temp. PR 1 (7359 741 9955 640 9904 8154 7569 3893 = 1) (else=0), gen(RECODE) drop temp. PR 1 • Time=118. 1 sec (Ouch! Much of the time is devoted to the destring command)

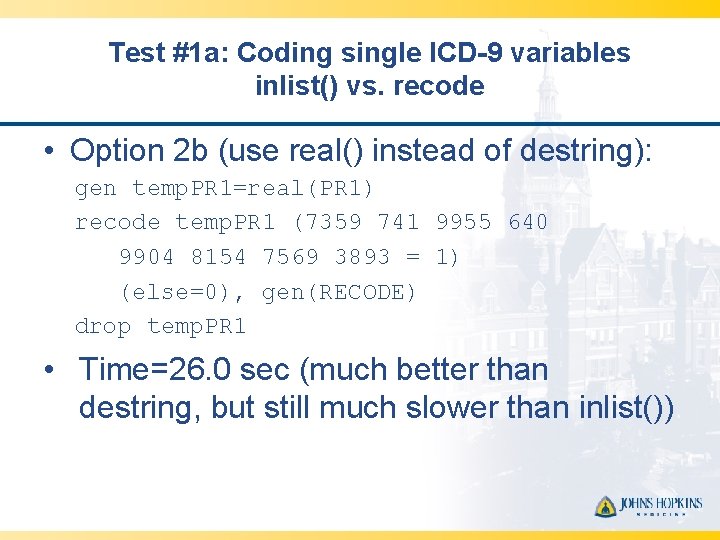
Test #1 a: Coding single ICD-9 variables inlist() vs. recode • Option 2 b (use real() instead of destring): gen temp. PR 1=real(PR 1) recode temp. PR 1 (7359 741 9955 640 9904 8154 7569 3893 = 1) (else=0), gen(RECODE) drop temp. PR 1 • Time=26. 0 sec (much better than destring, but still much slower than inlist())

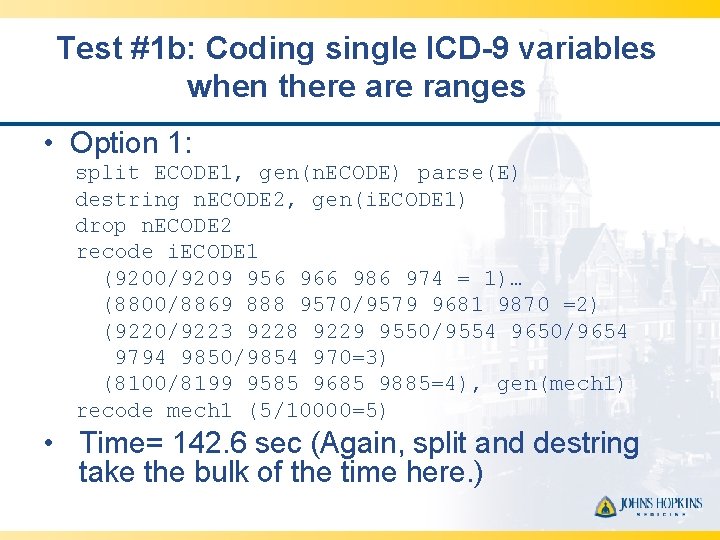
Test #1 b: Coding single ICD-9 variables when there are ranges • Option 1: split ECODE 1, gen(n. ECODE) parse(E) destring n. ECODE 2, gen(i. ECODE 1) drop n. ECODE 2 recode i. ECODE 1 (9200/9209 956 966 986 974 = 1)… (8800/8869 888 9570/9579 9681 9870 =2) (9220/9223 9228 9229 9550/9554 9650/9654 9794 9850/9854 970=3) (8100/8199 9585 9685 9885=4), gen(mech 1) recode mech 1 (5/10000=5) • Time= 142. 6 sec (Again, split and destring take the bulk of the time here. )

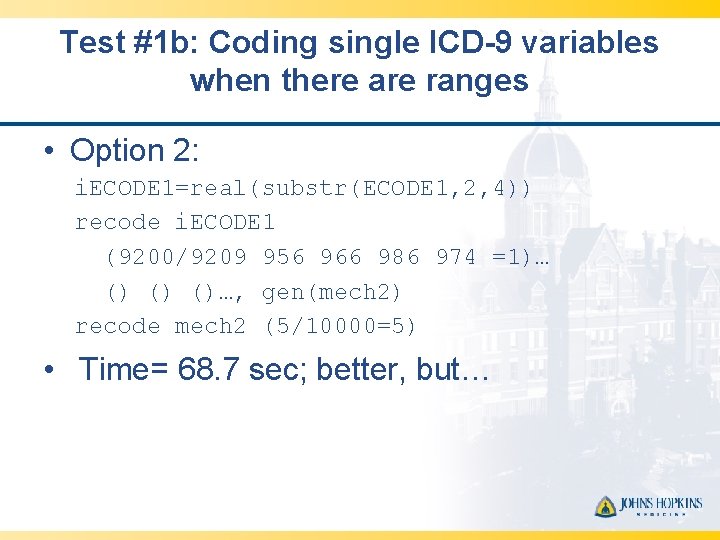
Test #1 b: Coding single ICD-9 variables when there are ranges • Option 2: i. ECODE 1=real(substr(ECODE 1, 2, 4)) recode i. ECODE 1 (9200/9209 956 966 986 974 =1)… () () ()…, gen(mech 2) recode mech 2 (5/10000=5) • Time= 68. 7 sec; better, but…

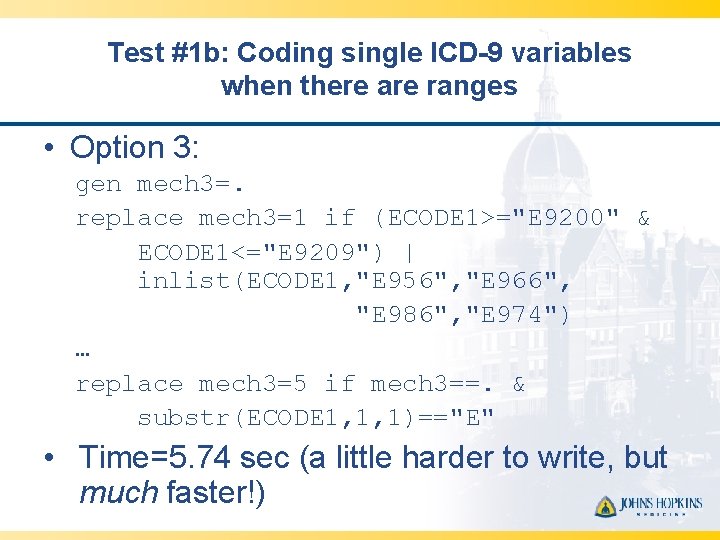
Test #1 b: Coding single ICD-9 variables when there are ranges • Option 3: gen mech 3=. replace mech 3=1 if (ECODE 1>="E 9200" & ECODE 1<="E 9209") | inlist(ECODE 1, "E 956", "E 966", "E 986", "E 974") … replace mech 3=5 if mech 3==. & substr(ECODE 1, 1, 1)=="E" • Time=5. 74 sec (a little harder to write, but much faster!)

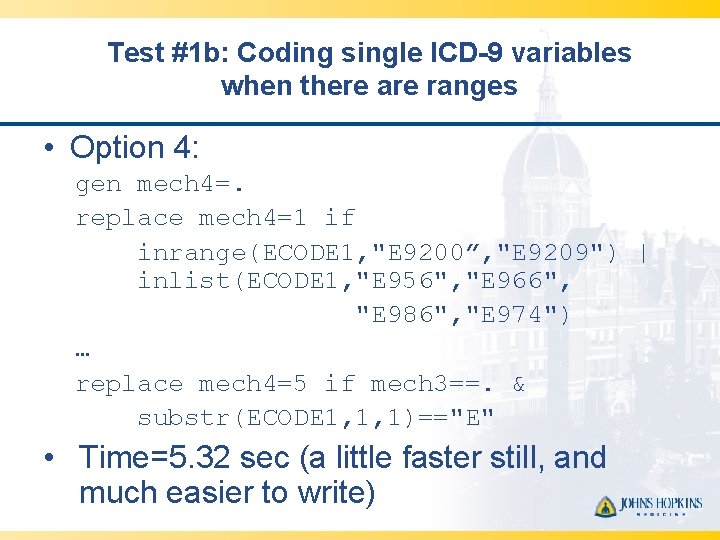
Test #1 b: Coding single ICD-9 variables when there are ranges • Option 4: gen mech 4=. replace mech 4=1 if inrange(ECODE 1, "E 9200”, "E 9209") | inlist(ECODE 1, "E 956", "E 966", "E 986", "E 974") … replace mech 4=5 if mech 3==. & substr(ECODE 1, 1, 1)=="E" • Time=5. 32 sec (a little faster still, and much easier to write)

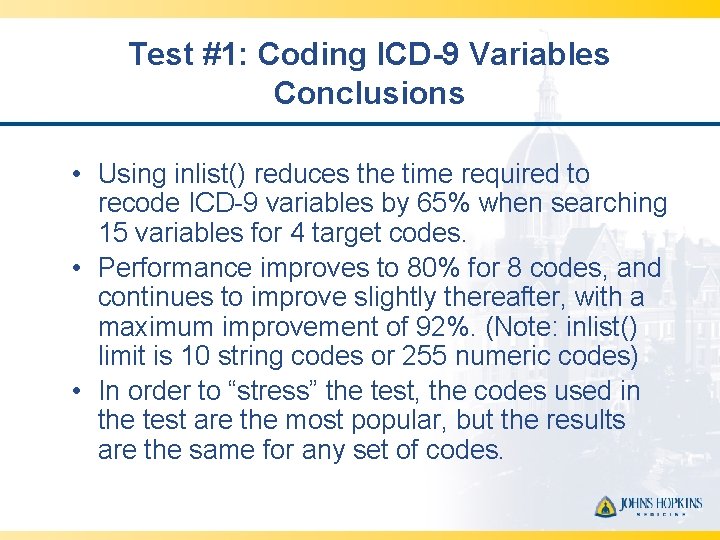
Test #1: Coding ICD-9 Variables Conclusions • Using inlist() reduces the time required to recode ICD-9 variables by 65% when searching 15 variables for 4 target codes. • Performance improves to 80% for 8 codes, and continues to improve slightly thereafter, with a maximum improvement of 92%. (Note: inlist() limit is 10 string codes or 255 numeric codes) • In order to “stress” the test, the codes used in the test are the most popular, but the results are the same for any set of codes.

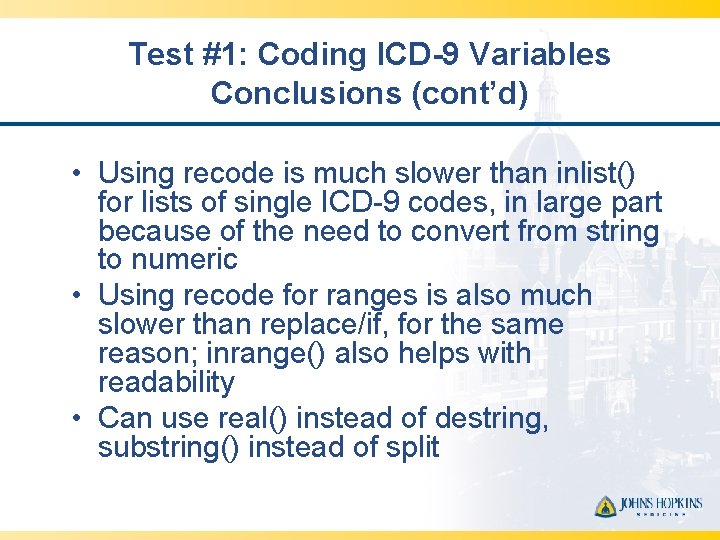
Test #1: Coding ICD-9 Variables Conclusions (cont’d) • Using recode is much slower than inlist() for lists of single ICD-9 codes, in large part because of the need to convert from string to numeric • Using recode for ranges is also much slower than replace/if, for the same reason; inrange() also helps with readability • Can use real() instead of destring, substring() instead of split

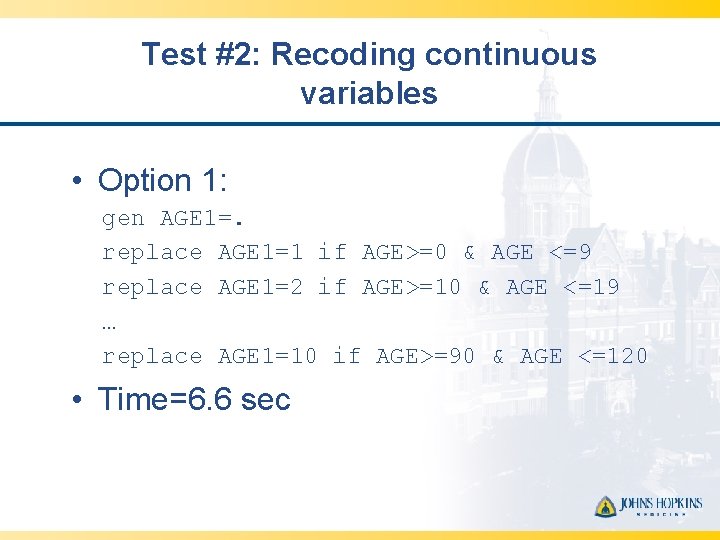
Test #2: Recoding continuous variables • Option 1: gen AGE 1=. replace AGE 1=1 if AGE>=0 & AGE <=9 replace AGE 1=2 if AGE>=10 & AGE <=19 … replace AGE 1=10 if AGE>=90 & AGE <=120 • Time=6. 6 sec

Test #2: Recoding continuous variables • Option 2: gen AGE 2=recode(AGE, 9, 19, 29, 39, 49, 59, 69, 79, 89, 120) • Time=0. 66 sec (exactly one-tenth of the time(!) and easier to write and read) • Caution: need to be careful with truly continuous variables that you are cutting at the right place

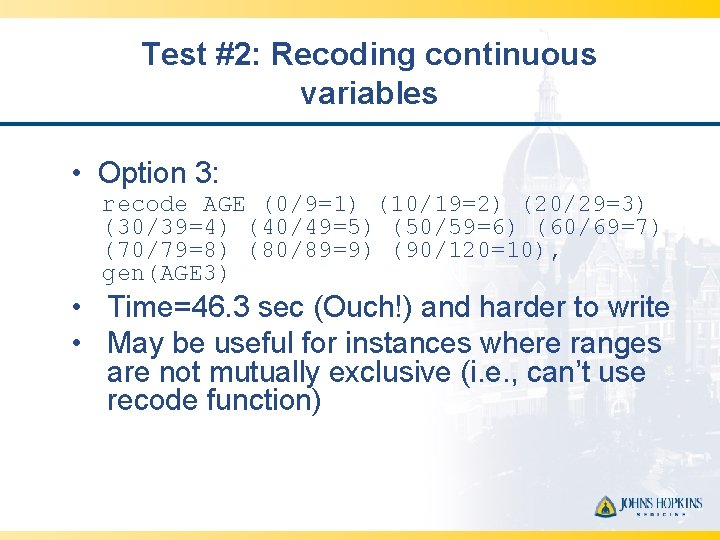
Test #2: Recoding continuous variables • Option 3: recode AGE (0/9=1) (10/19=2) (20/29=3) (30/39=4) (40/49=5) (50/59=6) (60/69=7) (70/79=8) (80/89=9) (90/120=10), gen(AGE 3) • Time=46. 3 sec (Ouch!) and harder to write • May be useful for instances where ranges are not mutually exclusive (i. e. , can’t use recode function)

Test #3: Reordering Values • Option 1: gen sex_new=sex replace sex_new=0 replace sex_new=5 replace sex_new=4 replace sex_new=1 replace sex_new=2 if if if sex_new==3 sex_new==2 sex_new==1 sex_new==5 sex_new==4 • Time=2. 0 sec; very cumbersome and hard to follow

Test #3: Reordering Values • Option 2: recode sex (3=0) (1=2) (2=1), gen(sex_new 1) • Time=15. 0 sec (Ouch! ); but, easier to write and MUCH easier to read) • Can also use recode to do things like: (3 4 = 0) // 3 and 4 are recoded to 0 (3/5 = 0) // 3, 4, and 5 are recoded to 0

Test #3: Reordering Values • Option 3: gen sex_new=sex replace sex_new=0 if sex==3 replace sex_new=1 if sex==2 replace sex_new=2 if sex==1 • Time=1. 4 sec (Faster than Option #1 by 40% and not too hard to read/write)

Test #4 De-stringing Numeric Values (e. g. , NSQIP age) • Option 1 (Variation of Test #3 Option #1): encode age, gen (age_new) replace age_new=180 if age_new==1 … replace age_new=900 if age_new==73 replace age_new=18 if age_new==180 … replace age_new=90 if age_new==900 • Time=25. 8 sec (NSQIP 2011; n=442, 149), • Always need to do “tab age_new, nolabel” because labels are messed up

Test #4 Destringing Numeric Values (e. g. , NSQIP age) • Option 2: destring age, gen(age_new 1) ignore(“+”) • Time=6. 3 sec (NSQIP 2011; n=442, 149); four times faster! • Caution: make sure it is clear that 89=89+

Test #4 a Removing Characters from ID Numbers (e. g. , XXX-XX-XXXX) • Option 1 destring SSN, ignore("-") gen(new. SSN 1) • Time=33. 0 sec

Test #4 a Removing Characters from ID Numbers (e. g. , XXX-XX-XXXX) • Option 2: gen long new. SSN 2= real(subinstr(SSN, "-", "", . )) • Time=1. 7 sec; almost 20 times faster! • Only useful if there a few characters to get rid of.

Future Tests • Confirm results for 10 years of NIS (about 80 million observations, nearly 50 Gb RAM) • Other Stata commands where there are multiple ways to do the same thing…any ideas? • Other programming practices found reviewing code written by colleagues and students

Implications • With 10 years of NIS, could save… – – 3 minutes per ICD-9 recode 1 minute per continuous variable categorization 6 seconds per variable reorder A lot more if you used recode • It all adds up! • Might make it less onerous to run recoding and cleaning programs more often instead of saving new copies of the dataset • Easier to read programs