Optimization of the Solid phase extraction column for










- Slides: 24

Optimization of the Solid phase extraction column for purificaion of DNA and RNA August 18 2009 Hussam Al. Muayad

Outline Brief history of the Progress in the lab Challenges: Standardizing chip design and getting a working model SPE Column efficiency put to question!!! Manipulating the column chemistry to increase the efficiency Near-Future work

Standardizing chip design and getting a working model 1. Guitar wires 2. Random chip dimensions and channel volume 3. Nanoport & epoxy May 2007

Brave attempts Major bonding problems at this stage Resin molds Embedded guitar wires (sepsis paper was completed using these very chips)


Features with high aspect ratio (up to 1. 5 mm feature height) More tolerance for deformation during boding Constant channel volume and constant SPE solution volume

Standardized Chip design and application Although not completely treated the problem of bonding lingers on with less hindrance to daily progress

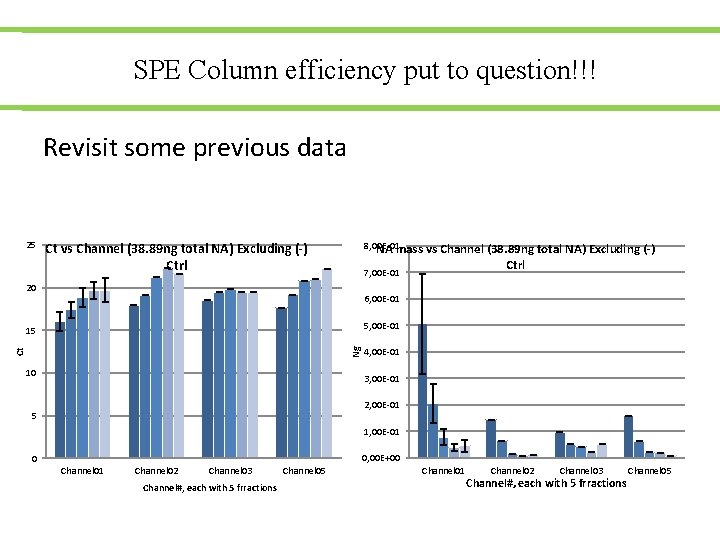
SPE Column efficiency put to question!!! Revisit some previous data 25 8, 00 E-01 NA mass vs Channel (38. 89 ng total NA) Excluding (-) Ctrl 7, 00 E-01 20 6, 00 E-01 5, 00 E-01 Ct Ng 15 10 4, 00 E-01 3, 00 E-01 2, 00 E-01 5 1, 00 E-01 0, 00 E+00 0 Channel 01 Channel 02 Channel 03 Channel#, each with 5 frractions Channel 05

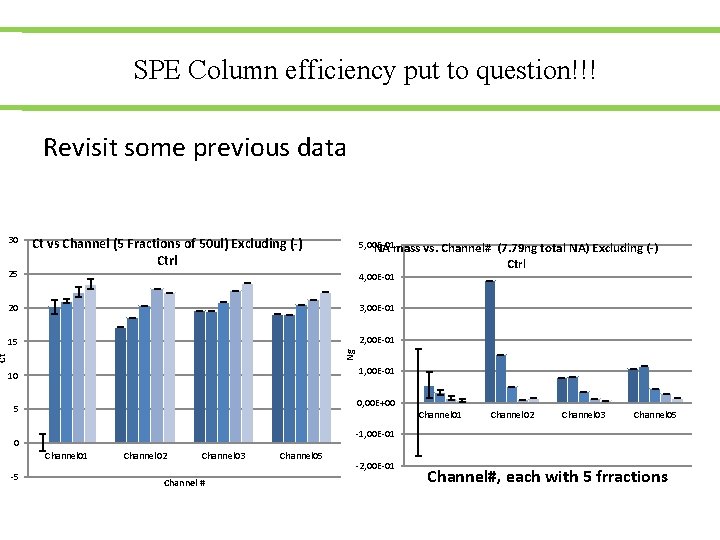
SPE Column efficiency put to question!!! Revisit some previous data 30 25 Ct vs Channel (5 Fractions of 50 ul) Excluding (-) Ctrl 5, 00 E-01 NA mass vs. Channel# 4, 00 E-01 3, 00 E-01 15 2, 00 E-01 Ct Ng 20 (7. 79 ng total NA) Excluding (-) Ctrl 1, 00 E-01 10 0, 00 E+00 5 Channel 01 Channel 03 Channel 05 -1, 00 E-01 0 Channel 01 -5 Channel 02 Channel 03 Channel # Channel 05 -2, 00 E-01 Channel#, each with 5 frractions

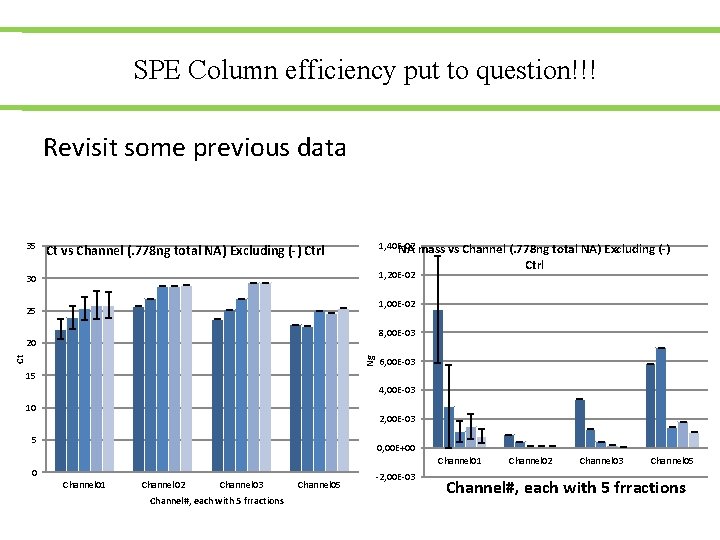
SPE Column efficiency put to question!!! Revisit some previous data 35 1, 40 E-02 NA mass vs Channel (. 778 ng total NA) Excluding (-) Ctrl 1, 20 E-02 30 1, 00 E-02 25 8, 00 E-03 Ng Ct 20 6, 00 E-03 15 4, 00 E-03 10 2, 00 E-03 5 0, 00 E+00 Channel 01 Channel 02 Channel 03 Channel#, each with 5 frractions Channel 05 -2, 00 E-03 Channel 02 Channel 03 Channel 05 Channel#, each with 5 frractions

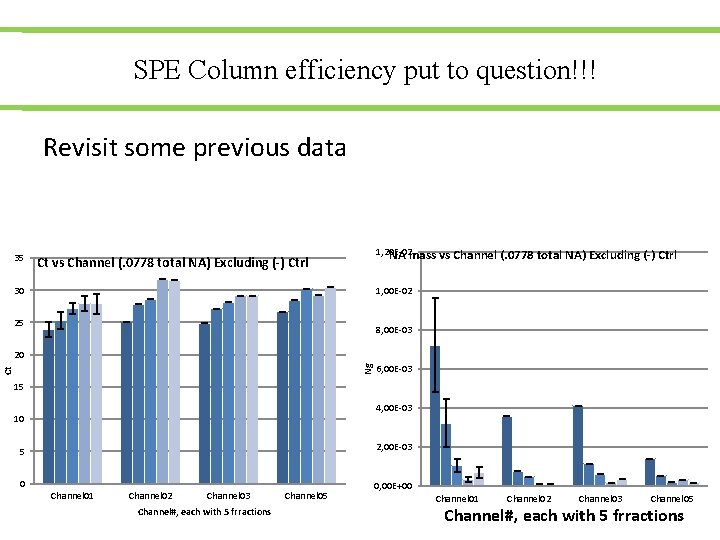
SPE Column efficiency put to question!!! Revisit some previous data 35 1, 20 E-02 NA mass vs Channel (. 0778 total NA) Excluding (-) Ctrl Ct vs Channel (. 0778 total NA) Excluding (-) Ctrl 30 1, 00 E-02 25 8, 00 E-03 Ct Ng 20 6, 00 E-03 15 4, 00 E-03 10 2, 00 E-03 5 0 Channel 01 Channel 02 Channel 03 Channel#, each with 5 frractions Channel 05 0, 00 E+00 Channel 01 Channel 02 Channel 03 Channel 05 Channel#, each with 5 frractions

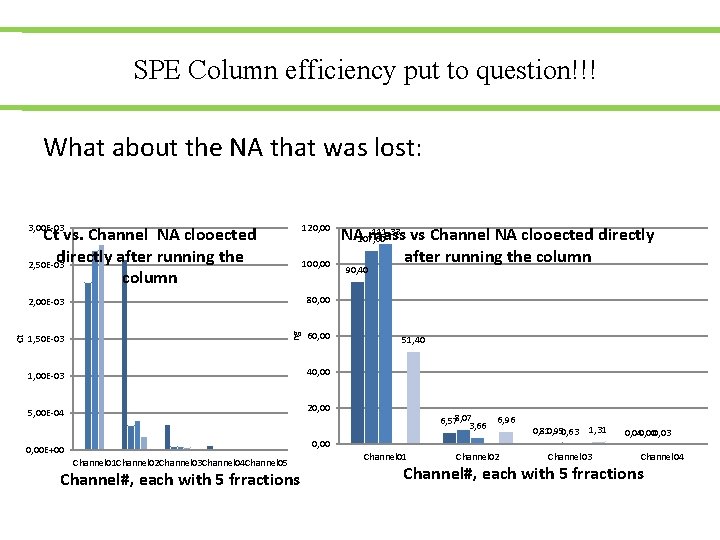
SPE Column efficiency put to question!!! What about the NA that was lost: 3, 00 E-03 120, 00 100, 00 2, 00 E-03 80, 00 1, 50 E-03 60, 00 ng Ct Ct vs. Channel NA clooected directly after running the 2, 50 E-03 column 1, 00 E-03 40, 00 5, 00 E-04 20, 00 111, 33 vs Channel NA clooected directly NA 107, 60 mass after running the column 90, 40 51, 40 6, 578, 07 3, 66 6, 96 0, 810, 950, 63 1, 31 0, 00 E+00 Channel 01 Channel 02 Channel 03 Channel 04 Channel 05 Channel#, each with 5 frractions Channel 01 Channel 02 Channel 03 0, 040, 000, 03 Channel 04 Channel#, each with 5 frractions

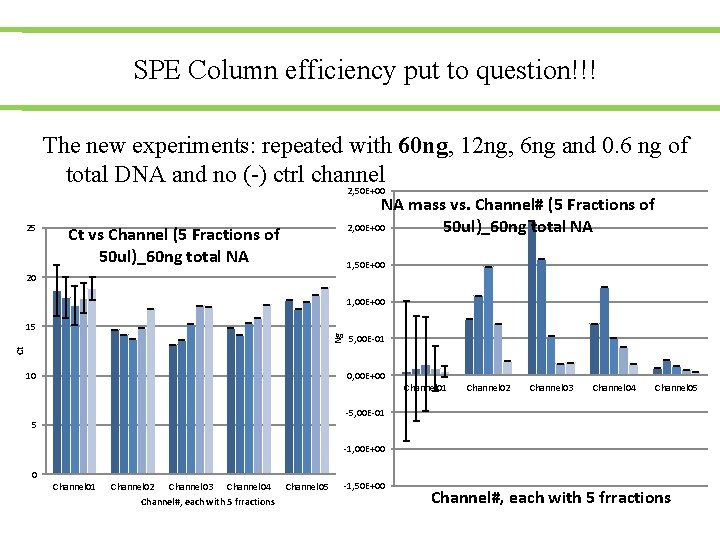
SPE Column efficiency put to question!!! The new experiments: repeated with 60 ng, 12 ng, 6 ng and 0. 6 ng of total DNA and no (-) ctrl channel 2, 50 E+00 25 NA mass vs. Channel# (5 Fractions of 2, 00 E+00 50 ul)_60 ng total NA Ct vs Channel (5 Fractions of 50 ul)_60 ng total NA 1, 50 E+00 20 1, 00 E+00 5, 00 E-01 Ct Ng 15 0, 00 E+00 10 Channel 01 Channel 02 Channel 03 Channel 04 Channel 05 -5, 00 E-01 5 -1, 00 E+00 0 Channel 01 Channel 02 Channel 03 Channel 04 Channel#, each with 5 frractions Channel 05 -1, 50 E+00 Channel#, each with 5 frractions

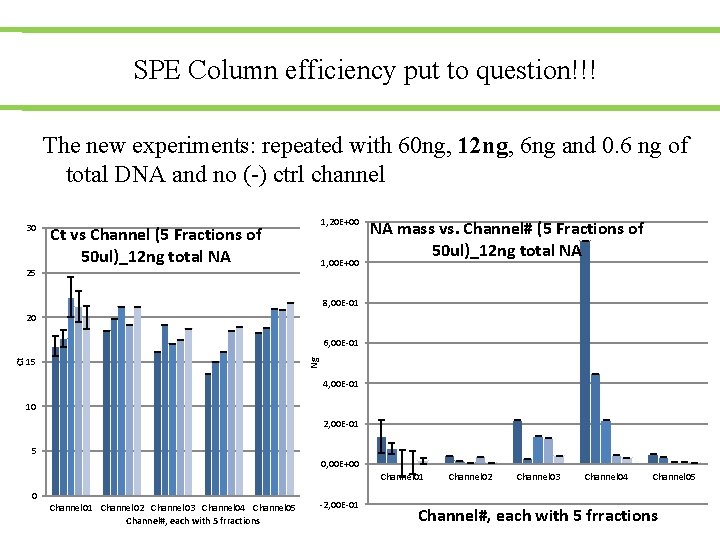
SPE Column efficiency put to question!!! The new experiments: repeated with 60 ng, 12 ng, 6 ng and 0. 6 ng of total DNA and no (-) ctrl channel 30 1, 20 E+00 Ct vs Channel (5 Fractions of 50 ul)_12 ng total NA 1, 00 E+00 25 NA mass vs. Channel# (5 Fractions of 50 ul)_12 ng total NA 8, 00 E-01 20 15 Ng Ct 6, 00 E-01 4, 00 E-01 10 2, 00 E-01 5 0, 00 E+00 Channel 01 Channel 02 Channel 03 Channel 04 Channel 05 Channel#, each with 5 frractions -2, 00 E-01 Channel 02 Channel 03 Channel 04 Channel 05 Channel#, each with 5 frractions

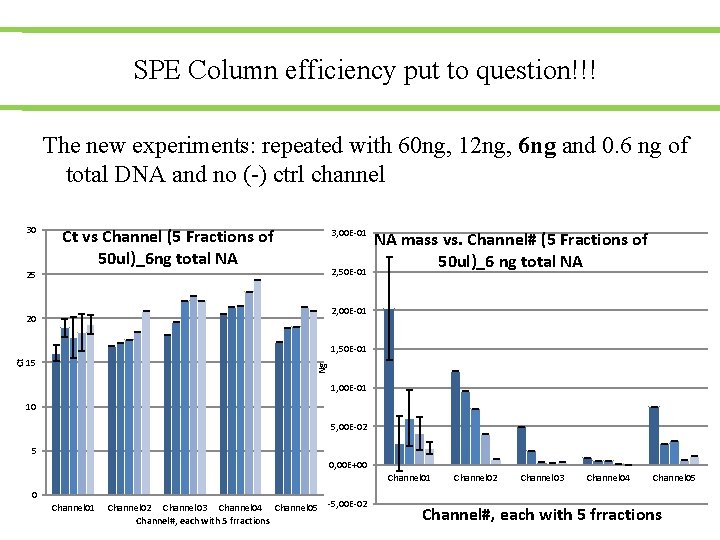
SPE Column efficiency put to question!!! The new experiments: repeated with 60 ng, 12 ng, 6 ng and 0. 6 ng of total DNA and no (-) ctrl channel 30 25 Ct vs Channel (5 Fractions of 50 ul)_6 ng total NA 3, 00 E-01 2, 50 E-01 NA mass vs. Channel# (5 Fractions of 50 ul)_6 ng total NA 2, 00 E-01 20 Ng Ct 1, 50 E-01 15 1, 00 E-01 10 5, 00 E-02 5 0, 00 E+00 Channel 01 Channel 02 Channel 03 Channel 04 Channel 05 -5, 00 E-02 Channel#, each with 5 frractions Channel 02 Channel 03 Channel 04 Channel 05 Channel#, each with 5 frractions

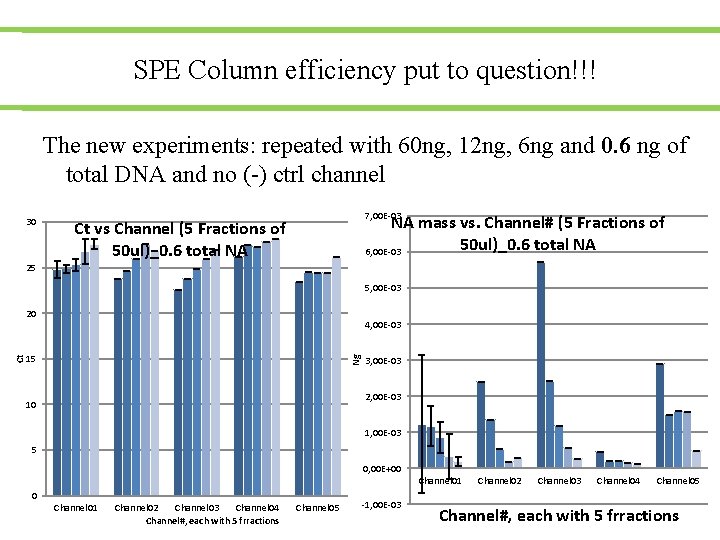
SPE Column efficiency put to question!!! The new experiments: repeated with 60 ng, 12 ng, 6 ng and 0. 6 ng of total DNA and no (-) ctrl channel 30 7, 00 E-03 NA mass vs. Channel# (5 Fractions of 50 ul)_0. 6 total NA 6, 00 E-03 Ct vs Channel (5 Fractions of 50 ul)_0. 6 total NA 25 5, 00 E-03 4, 00 E-03 15 Ng Ct 20 3, 00 E-03 2, 00 E-03 10 1, 00 E-03 5 0, 00 E+00 Channel 01 Channel 02 Channel 03 Channel 04 Channel#, each with 5 frractions Channel 05 -1, 00 E-03 Channel 02 Channel 03 Channel 04 Channel 05 Channel#, each with 5 frractions

SPE Column efficiency put to question!!! Comparison between 7. 78 ng and 6 ng total NA 5, 00 E-01 NA mass vs. Channel# 6, 00 E-01 (7. 79 ng total NA) Excluding (-) Ctrl 5, 00 E-01 NA mass vs. Channel# (5 Fractions of 50 ul)_6 ng total NA 4, 00 E-01 3, 00 E-01 Ng 3, 00 E-01 2, 00 E-01 1, 00 E-01 0, 00 E+00 Channel 01 -1, 00 E-01 Channel 02 Channel 03 Channel 05 Channel#, each with 5 frractions 0, 00 E+00 Channel 01 -1, 00 E-01 Channel 02 Channel 03 Channel 04 Channel 05 Channel#, each with 5 frractions

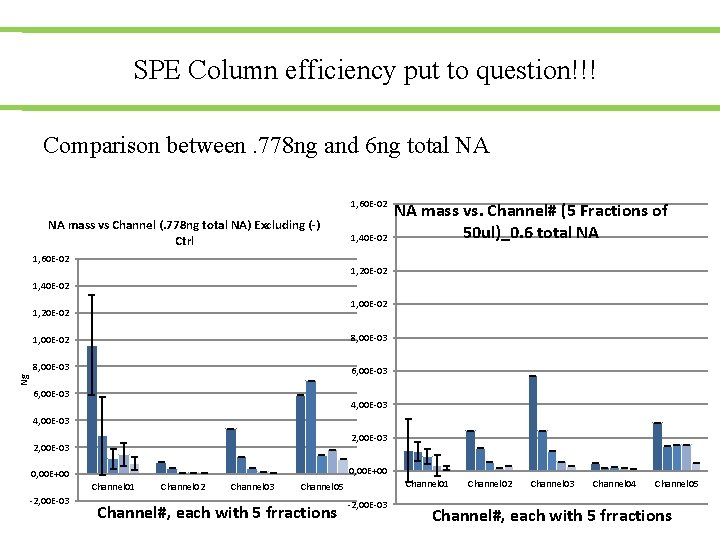
SPE Column efficiency put to question!!! Comparison between. 778 ng and 6 ng total NA 1, 60 E-02 NA mass vs Channel (. 778 ng total NA) Excluding (-) Ctrl 1, 60 E-02 1, 40 E-02 NA mass vs. Channel# (5 Fractions of 50 ul)_0. 6 total NA 1, 20 E-02 1, 40 E-02 1, 00 E-02 1, 20 E-02 8, 00 E-03 6, 00 E-03 Ng 1, 00 E-02 6, 00 E-03 4, 00 E-03 2, 00 E-03 0, 00 E+00 Channel 01 -2, 00 E-03 Channel 02 Channel 03 Channel 01 Channel 05 Channel#, each with 5 frractions -2, 00 E-03 Channel 02 Channel 03 Channel 04 Channel 05 Channel#, each with 5 frractions

Manipulating the column chemistry to increase the efficiency Many variables to be considered 1. Volume of the Column, depends on channel dimension an design 2. The packing of silica , 1 x, 2 x… 10 x. . . 100 x (1 x being 100µl silica solution against 100µl polymer solution) 3. The diameter of the silica spheres (700 nm, 3 um…. . )… source the types 4. The percentages of polymers in the SPE mix (40%BUMA +60%EMDA, 60%BUMA+40%EDMA) 5. Duration of UV crosslinking 6. Additional important factors like flow rate or incubation time.

Manipulating the column chemistry to increase the efficiency 4µl, 3. 0µm silica Regular channel 4µl, 700 nm silica 13µl, 700 nm 2 X silica 13µl, 3. 0µm silica 13µl, 700 nm silica The Following experiment was carried out using the standard procedure • 60 ul of DNA was run into one set • 50 ul of RNA was run into a second set

PCR results 4µl, 3. 0µm silica Regular channel – Jessie found out that the RNA stock she has didn’t produce a proper standard curve which indicates the disintegration of the RNA 4µl, 700 nm silica 13µl, 700 nm 2 X silica 13µl, 3. 0µm silica 13µl, 700 nm silica – AB RT-PCR machine crashed while doing this PCR this morning with the DNA chip – Results of these chips are to follow shortly

Near-Future work in order or importance • Follow a systemic method to manipulate the SPE column based on the literature and calculate a close approximation of the NA that should be captured by the silica This includes as a first step, testing multiple diameters of silica spheres and different volumes of silica The second step in case the first fails, is to change/modify chemical composition of channel Important question to answer: what is the saturation threshold of the SPE per unit of volume • Assuming results of the lambda phage are satisfactory it’s time to move to the testing of purified bacterial DNA and/or E. coli • Simultaneously work on a simple channel design (with minor modification like the one displayed) to incorporate a larger volume of silica and perhaps increase the interaction interface between the fluids and the SPE

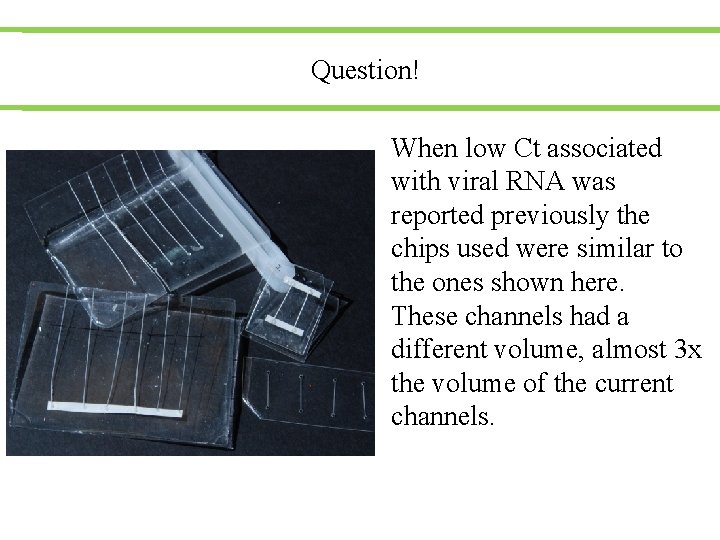
Question! When low Ct associated with viral RNA was reported previously the chips used were similar to the ones shown here. These channels had a different volume, almost 3 x the volume of the current channels.

Thank You!