Nucleotides Nucleic Acids and Heredity Bettelheim Brown Campbell

Nucleotides, Nucleic Acids and Heredity Bettelheim, Brown, Campbell and Farrell Chapter 25—part 2
DNA and RNA • The three differences in structure between DNA and RNA are – DNA bases are A, G, C, and T; the RNA bases are A, G, C, and U – the sugar in DNA is 2 -deoxy-D-ribose; 2 -deoxy-D-ribose in RNA it is D-ribose – DNA is always double stranded; stranded there are several kinds of RNA, all of which are singlestranded
RNA • RNA molecules are classified according to their structure and function
Other RNAs • RNA molecules are classified according to their structure and function • sn. RNA –small nuclear RNA (100 -200 b) Combine to make sn. RNPs to help processing of m. RNA for export from nucleus • mi. RNA—micro. RNA • Bind to m. RNA in development • si. RNA—small interfering RNA • Knock out m. RNA for genes that are undesirable
t-RNA Structure • Contains some modified nucleotides, such as 1 -methylguanosine • Actual 3 -D shape is an L • Often shown as a 2 -D “clover leaf” shape with three “loops” • Some H-bonding between bases at base of loops • Anticodon bases contained on middle loop • 3’ end has CCA as terminal bases • 3’ end carries the amino acid

“Clover Leaf” Structure of t. RNA
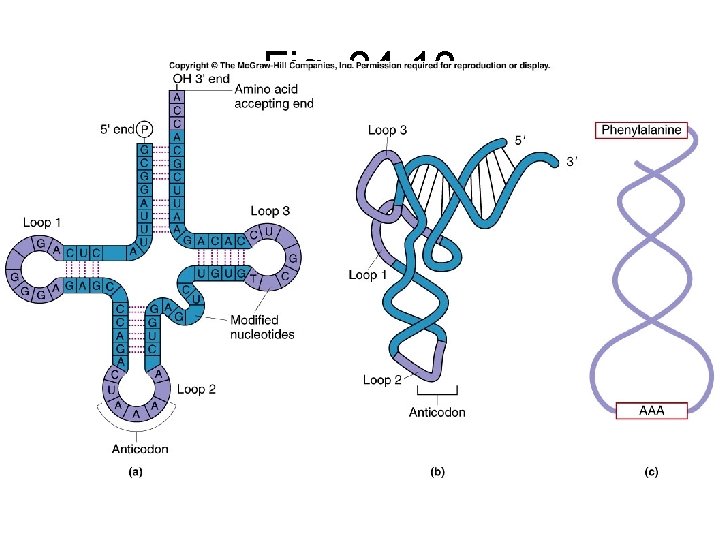
Fig. 24. 10

Nucleic Acids and Heredity • • Chromosomes exist in pairs (23 pairs in humans) Inherit one DNA copy from each parent. Most cells in our body contain copies of both Genetic information is carried in the sequence of bases along the DNA strands. • Information is passed to daughter cells when cell divides.
Genes, Exons, and Introns • Gene: a segment of DNA that carries a base sequence that directs the synthesis of a particular protein, t. RNA, or m. RNA – there are many genes in one DNA molecule – in bacteria the gene is continuous – in higher organisms the gene is discontinuous • Exon: a section of DNA that, when transcribed, codes for a protein or RNA • Intron: a section of DNA that does not code for anything functional
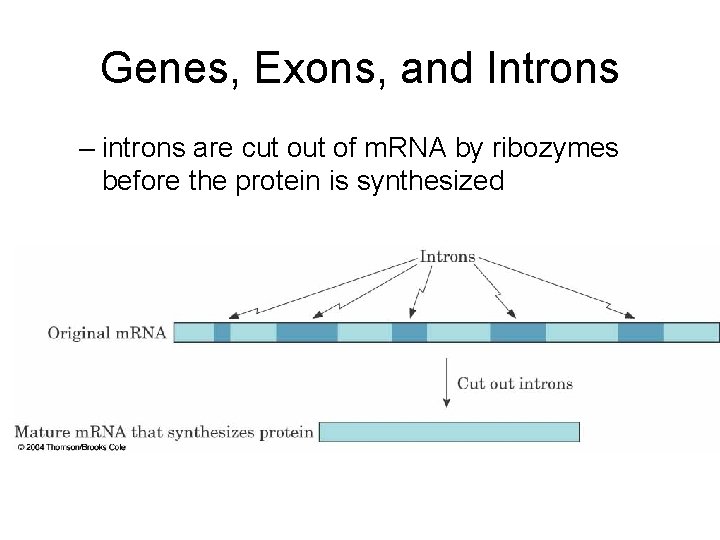
Genes, Exons, and Introns – introns are cut of m. RNA by ribozymes before the protein is synthesized
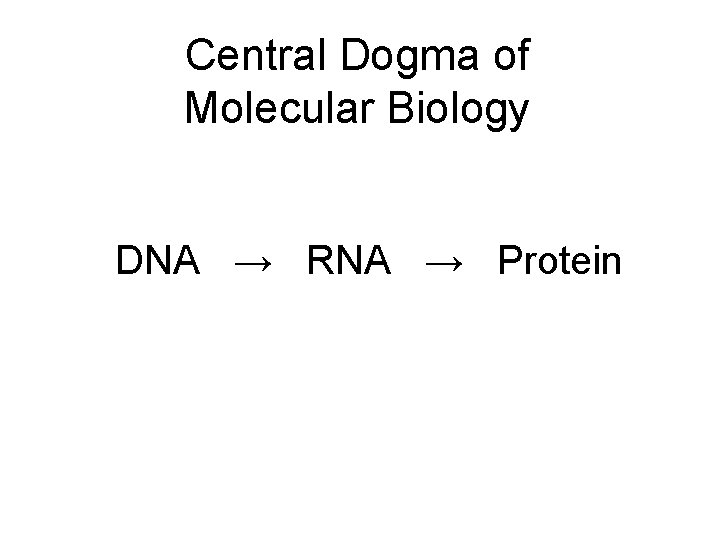
Central Dogma of Molecular Biology DNA → RNA → Protein
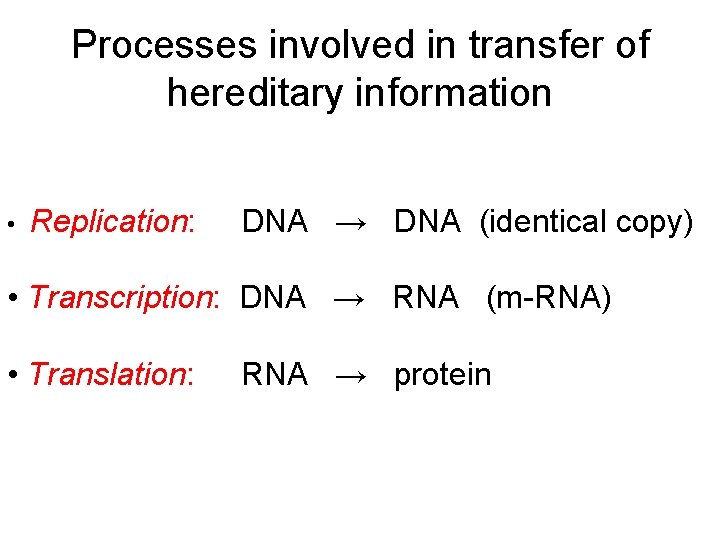
Processes involved in transfer of hereditary information • Replication: DNA → DNA (identical copy) • Transcription: DNA → RNA (m-RNA) • Translation: RNA → protein
DNA Replication • Replication involves separation of the two original strands and synthesis of two new daughter strands using the original strands as templates – DNA double helix unwinds at a specific point called an origin of replication – DNA replication is bidirectional: bidirectional chains are synthesized in both directions from the origin of replication – At each origin of replication, there are two replication forks where new polynucleotide strands are formed
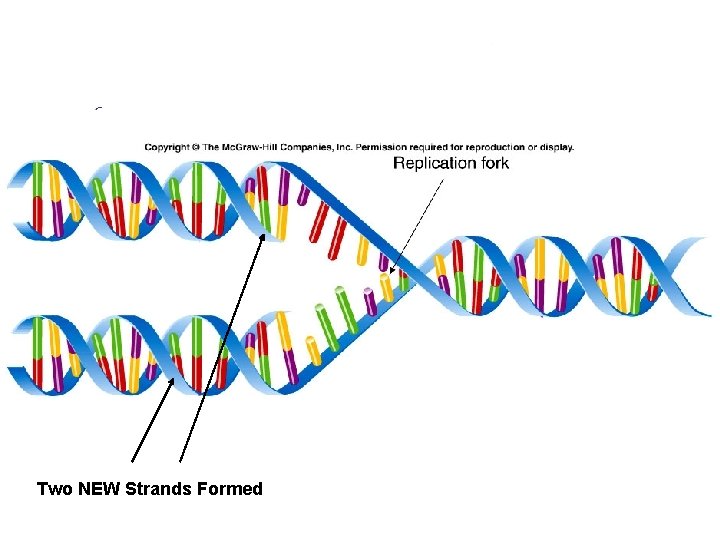
Two NEW Strands Formed

Steps in Replication 1. Weaken DNA-Histone interactions – Histone Acetylase interferes with +/- interaction by adding acetyl group to lysine amino groups 2. Relax higher DNA superstructure – Topoisomerases (gyrases) eliminate supercoiling of DNA by binding to one strand (via tyrosine and phosphate bond), nicking DNA, uncoiling DNA and rejoining DNA segments 3. Unwind Double Helix – Helicases attach to one strand separate the two strands (uses ATP for energy)
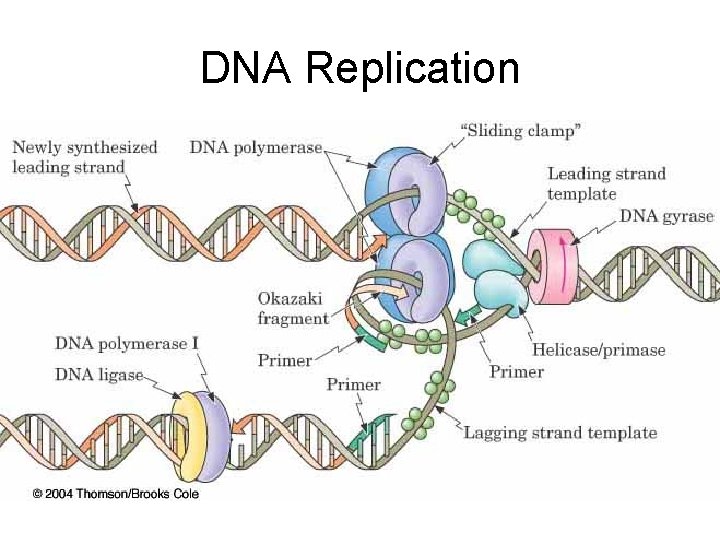
DNA Replication

Steps in Replication 4. Primer/Primases – Short RNA sequences needed to start DNA synthesis – Catalyzed by Primase 5. Polymerization (actual synthesis of new DNA strands) – – DNA Polymerase Catalyzes attachment base to new strand New base is complementary to template base Short Okasaki fragments formed (ca 200 bases) 6. Ligation – DNA ligase joins Okasaki fragments
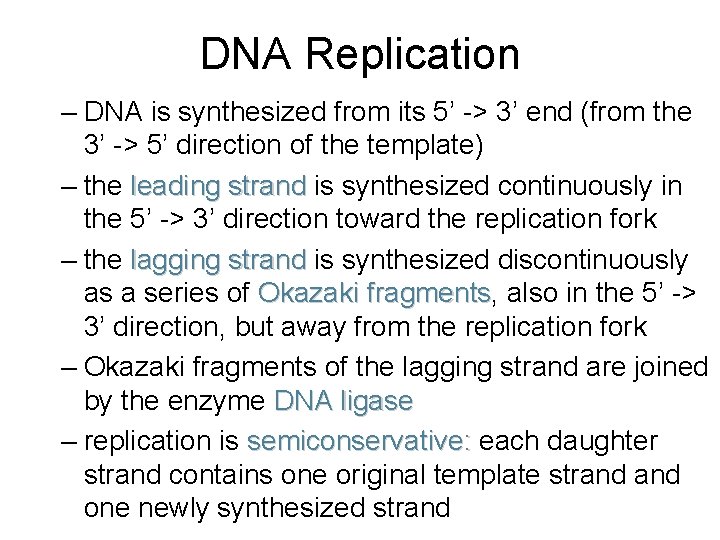
DNA Replication – DNA is synthesized from its 5’ -> 3’ end (from the 3’ -> 5’ direction of the template) – the leading strand is synthesized continuously in the 5’ -> 3’ direction toward the replication fork – the lagging strand is synthesized discontinuously as a series of Okazaki fragments, fragments also in the 5’ -> 3’ direction, but away from the replication fork – Okazaki fragments of the lagging strand are joined by the enzyme DNA ligase – replication is semiconservative: each daughter strand contains one original template strand one newly synthesized strand
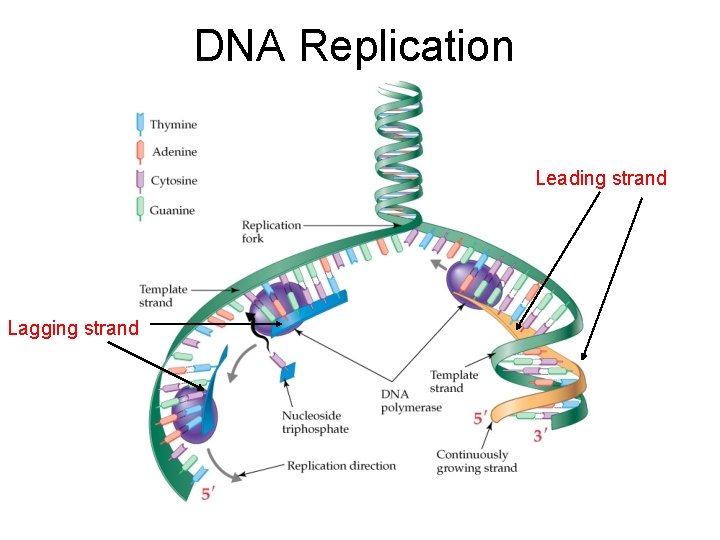
DNA Replication Leading strand Lagging strand
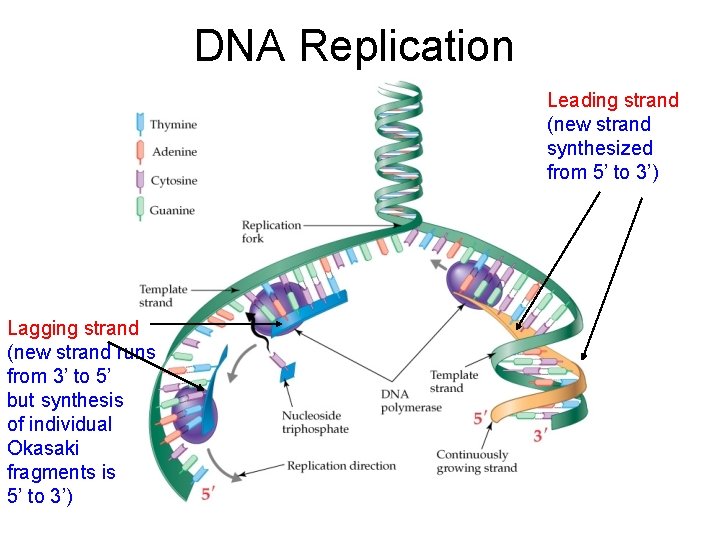
DNA Replication Leading strand (new strand synthesized from 5’ to 3’) Lagging strand (new strand runs from 3’ to 5’ but synthesis of individual Okasaki fragments is 5’ to 3’)
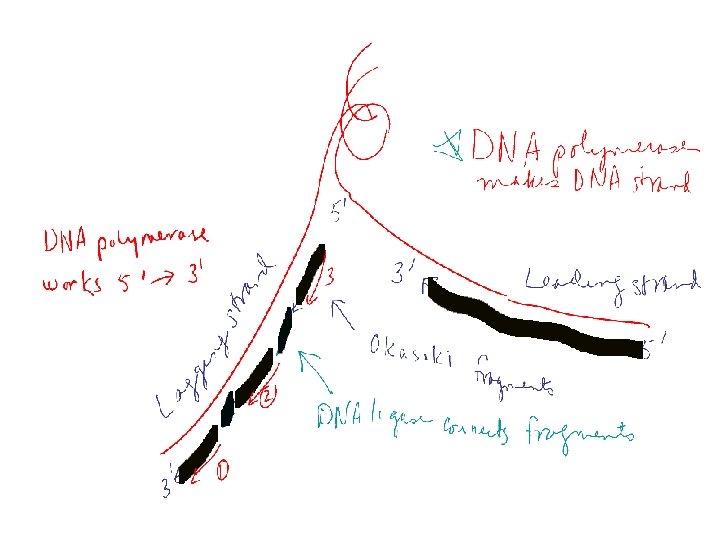
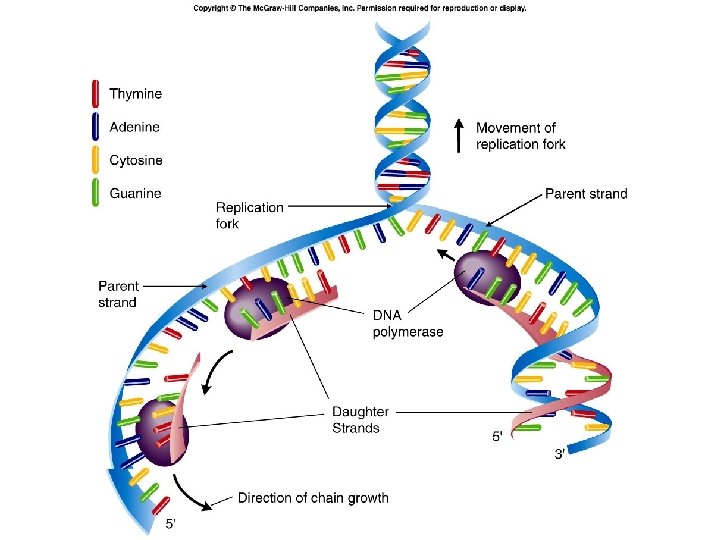
Fig. 24. 9
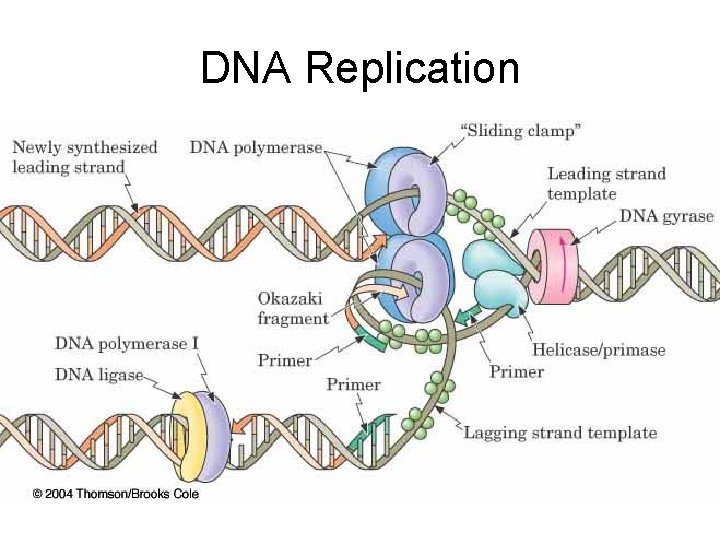
DNA Replication
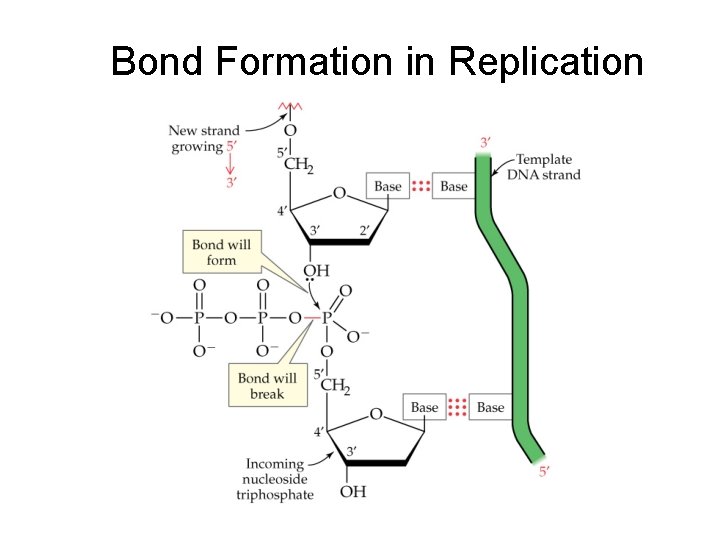
Bond Formation in Replication

DNA Replication Old strand: New strand: ATTCGTAAAGGTC TAAGCATT

DNA Repair • Cells have DNA repair enzymes that can detect, recognize, and repair mutations in DNA • Base excision repair (BER): (BER) one of the most common repair mechanisms – DNA glycosylase recognizes the “damaged” base and cuts out the base, leaving the sugar-phosphate backbone AP (apurinic or apyrimidinic) site created ap ap

DNA Repair • Endonuclease Endonucleas catalyzes the hydrolysis of the backbone – an exonuclease liberates the sugar-phosphate unit of the damaged site – DNA polymerase inserts the correct nucleotide – DNA ligase seals the backbone to complete the repair

DNA Repair • NER (nucleotide excision repair) removes and repairs up to 24 -32 units by a similar mechanism involving a number of repair enzymes
- Slides: 28