Nucleosome Positioning Transcription Factor Identification ZHANG ZHIZHUO MAY














- Slides: 30

Nucleosome Positioning & Transcription Factor Identification ZHANG ZHIZHUO MAY 2010

Outline �Basic Concept �Nucleosome Positioning and Gene Regulation General Transcription Mechanism Genomic Organization of Nucleosomes The Organization of Nucleosomes on Genes Control of DNA Access �Nucleosome Related TF Identification Enhancer Identification Integrating Histone Information to Predict TF binding sites

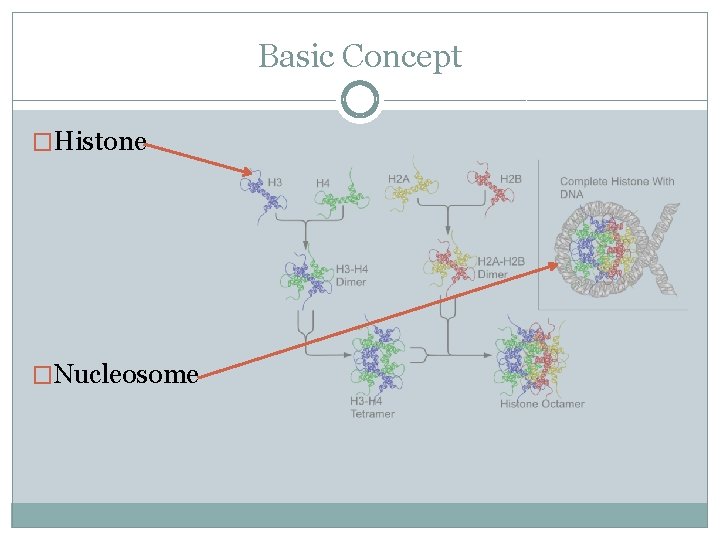
Basic Concept �Histone �Nucleosome

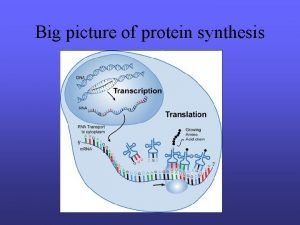
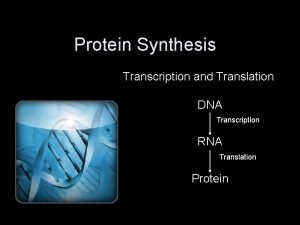
Basic Concept �Histone �Nucleosome �Linker-DNA between two nucleosomes �RNA Poly. II An enzyme catalyzes the transcription of DNA �TBP TATA binding protein and a necessary component of RNA polymerase

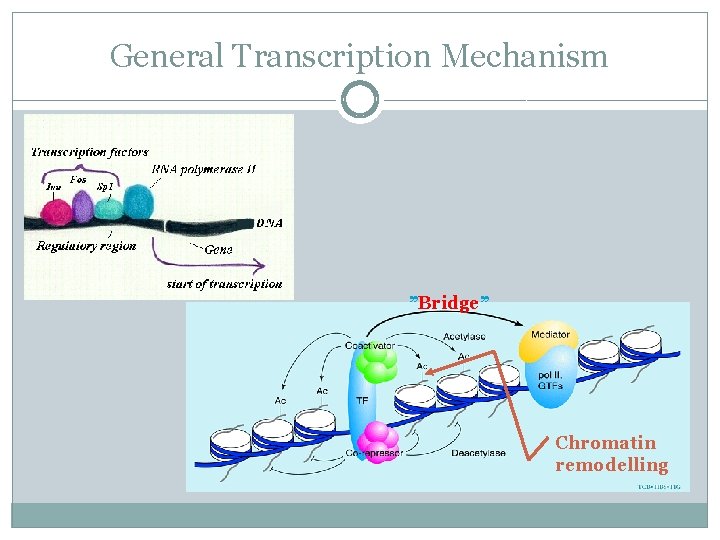
General Transcription Mechanism ”Bridge” Chromatin remodelling

General Transcription Mechanism

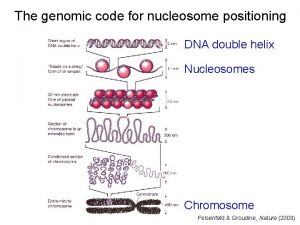
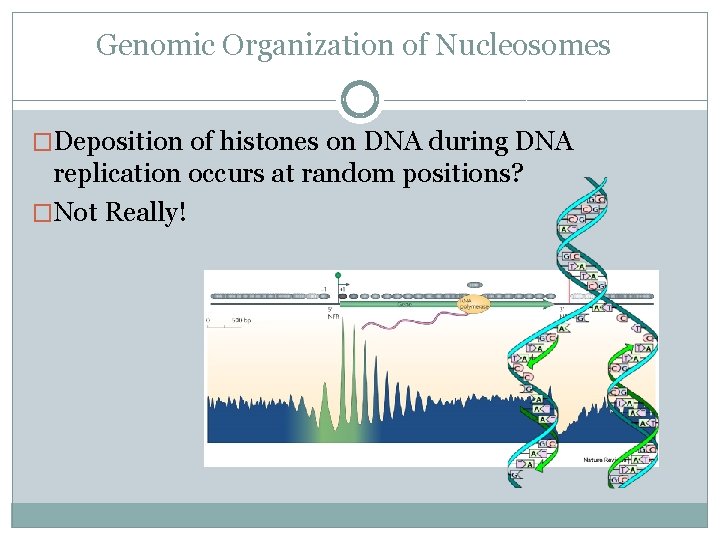
Genomic Organization of Nucleosomes �Deposition of histones on DNA during DNA replication occurs at random positions? �Not Really!

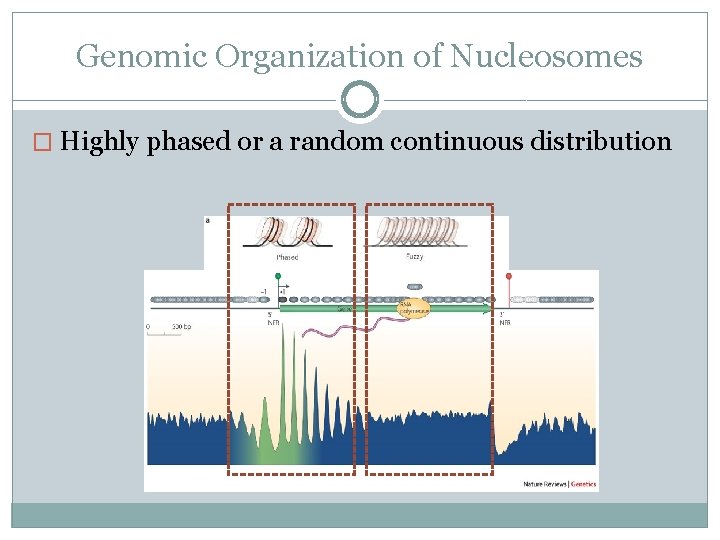
Genomic Organization of Nucleosomes � Highly phased or a random continuous distribution

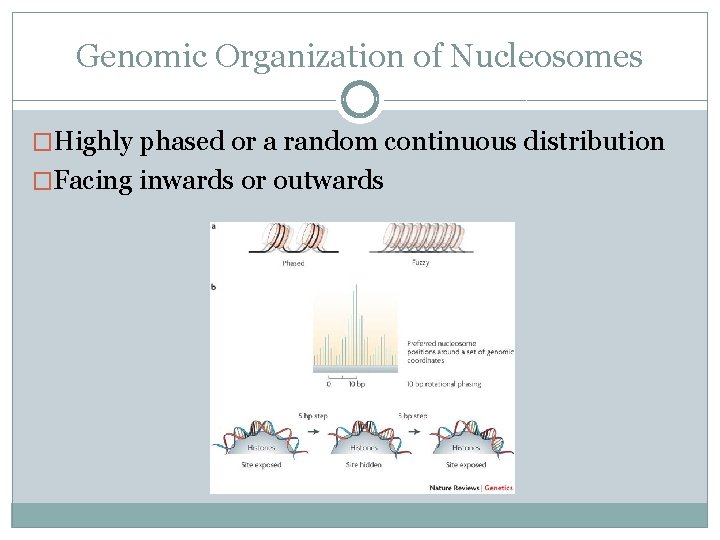
Genomic Organization of Nucleosomes �Highly phased or a random continuous distribution �Facing inwards or outwards

Distance between Positioned nucleosomes �Tend to be fixed distance �short stretches of linker DNA 165 bp (18 bp linker) in S. cerevisiae 175 bp (28 bp linker) in D. melanogaste 185 bp (38 bp linker) in humans �ISWI complex models the spacing �Linkers might have regulatory functions

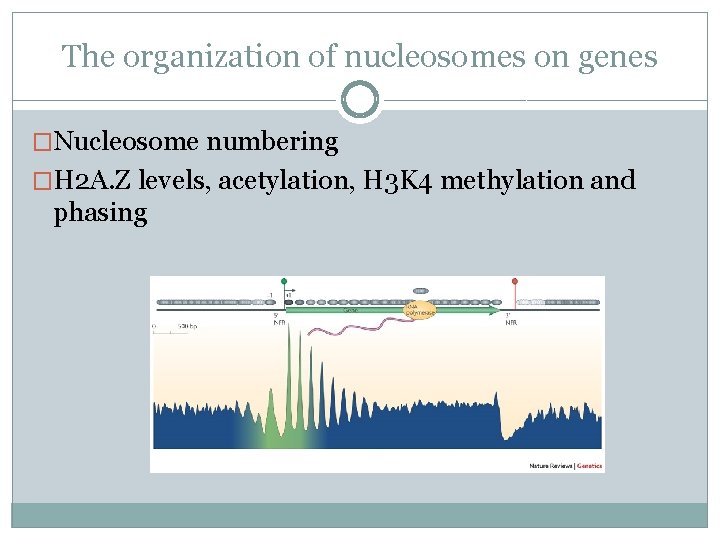
The organization of nucleosomes on genes �Nucleosome numbering

The organization of nucleosomes on genes �Nucleosome numbering �H 2 A. Z levels, acetylation, H 3 K 4 methylation and phasing

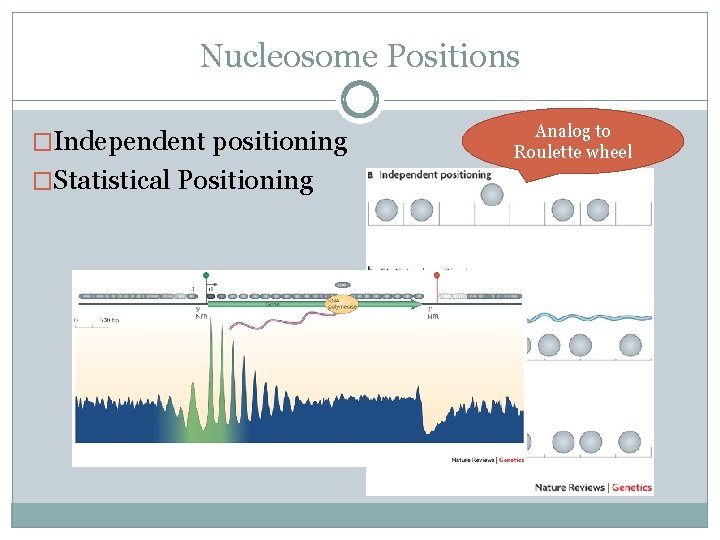
Nucleosome Positions �Independent positioning �Statistical Positioning Analog to Roulette wheel

Nucleosome-Free Region(NFR) �Poly(d. A: d. T) tracts contribute to rigidity �We thought promoter regions would be occluded by nucleosomes except when they were activated. �But in fact, NFRs demonstrated that open promoter states are stable and common, even at genes that are transcribed so infrequently.

Nucleosome-Free Region(NFR) �Low basal levels of leaky transcription might have a general housekeeping function �Open architecture of the 5' NFR is necessary for the initial 'pioneering' polymerase or whether transcription itself establishes the NFR from the closed state (after the last transcription)

Transcription start site selection by nucleosomes? � Most promoters seem to lack core promoter elements, including a TATA box, the TFIIB recognition element (BRE), INR, downstream promoter element (DPE) or motif ten element (MTE)

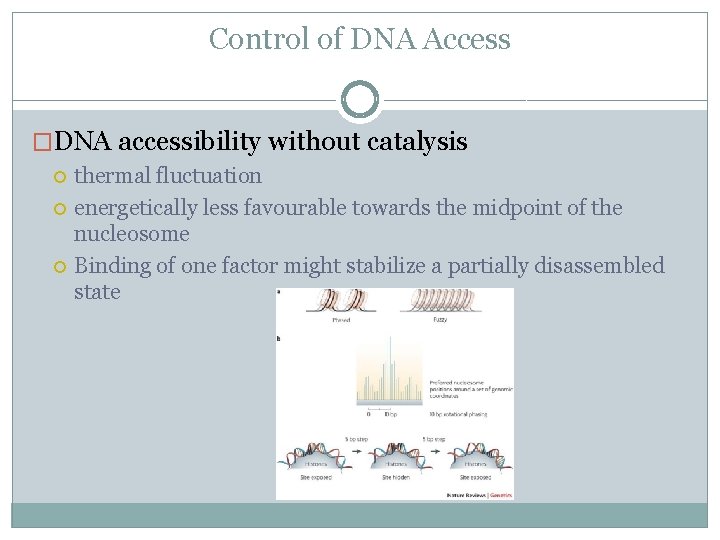
Control of DNA Access �DNA accessibility without catalysis thermal fluctuation energetically less favourable towards the midpoint of the nucleosome Binding of one factor might stabilize a partially disassembled state

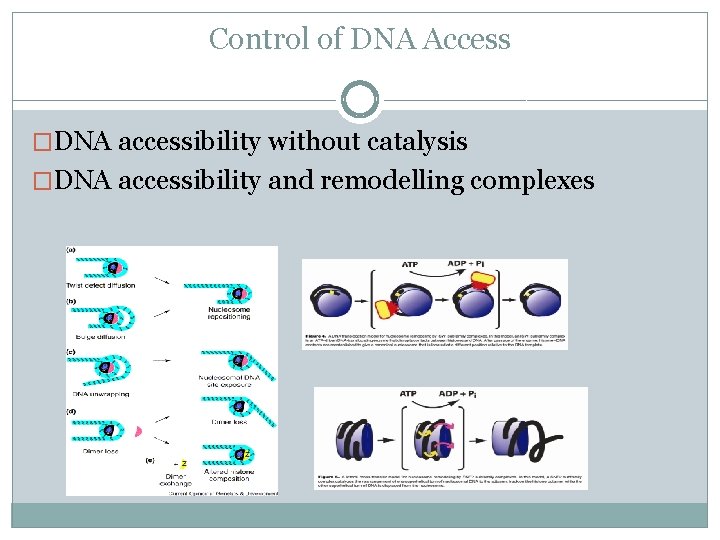
Control of DNA Access �DNA accessibility without catalysis �DNA accessibility and remodelling complexes

Control of DNA Access �DNA accessibility without catalysis �DNA accessibility and remodelling complexes �Nucleosome eviction

Nucleosome Related TF Identification Nucleosome dynamics define transcriptional enhancers. Nat Genet 42: 343 -347. Genome-wide prediction of transcription factor binding sites using an integrated model. Genome Biol 11: R 7.

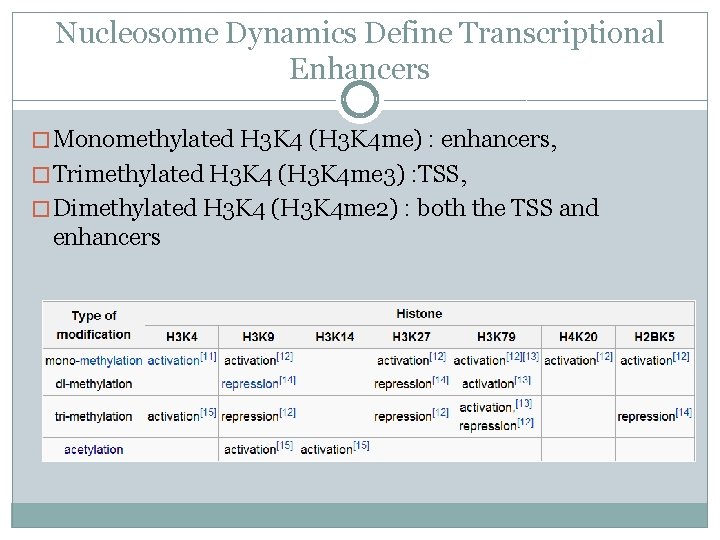
Nucleosome Dynamics Define Transcriptional Enhancers � Monomethylated H 3 K 4 (H 3 K 4 me) : enhancers, � Trimethylated H 3 K 4 (H 3 K 4 me 3) : TSS, � Dimethylated H 3 K 4 (H 3 K 4 me 2) : both the TSS and enhancers

H 3 K 4 me 2 are destabilized at AR binding site, but better positioning at flanking loci. FOXA 1 as pioneer factor to stabilize the nucleosomes.

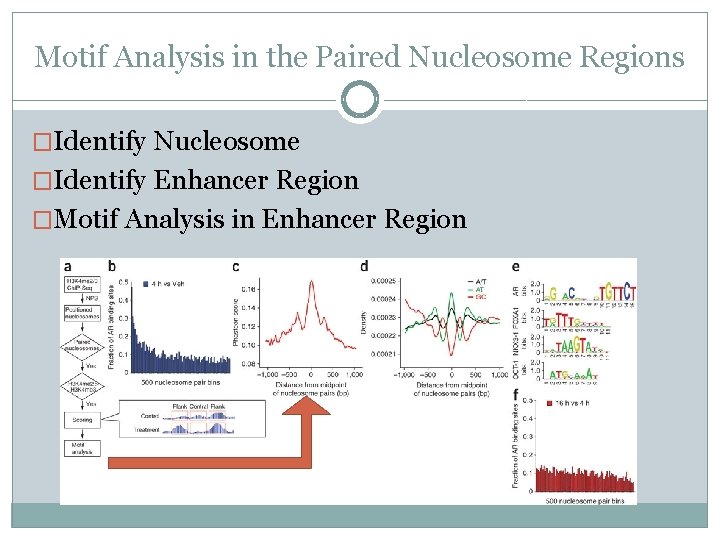
Motif Analysis in the Paired Nucleosome Regions �Identify Nucleosome

Motif Analysis in the Paired Nucleosome Regions �Identify Nucleosome �Identify Enhancer Region

Motif Analysis in the Paired Nucleosome Regions �Identify Nucleosome �Identify Enhancer Region �Motif Analysis in Enhancer Region

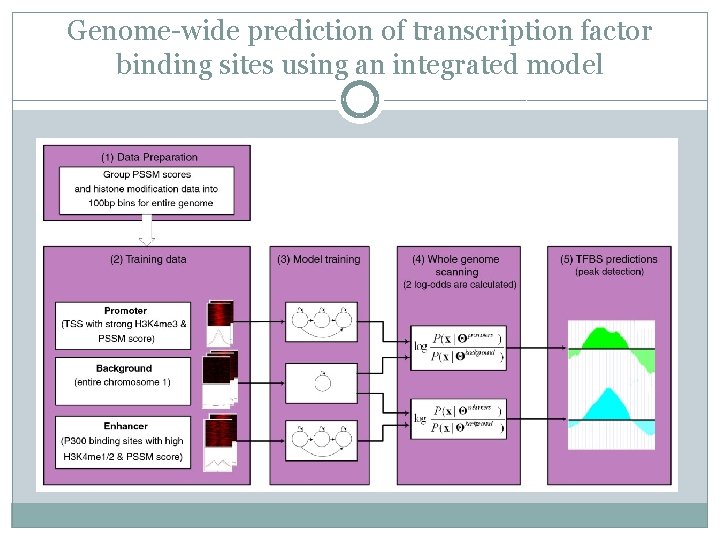
Genome-wide prediction of transcription factor binding sites using an integrated model � Eight chromatin marks (H 3, H 3 K 4 me 1, H 3 K 4 me 2, H 3 K 4 me 3, H 3 K 9 me 3, H 3 K 36 me 3, H 3 K 20 me 3, and H 3 K 27 me 3)

Genome-wide prediction of transcription factor binding sites using an integrated model

Future work �Finding candidate nucleosome organizing factors �Better understand nucleosome positioning

Reference � I) NUCLEOSOME POSITIONING. II) NUCLEOSOME REMODELING. BCH 6415. http: //www. med. ufl. edu/biochem/tyang/ � http: //www. uio. no/studier/emner/matnat/molbio/MBV 4230/ � Nucleosome dynamics define transcriptional enhancers, Nature genetics 42 (4): 343 -7, 2010 Apr � Nucleosome positioning and gene regulation: advances through genomics, Nature reviews. Genetics 10 (3): 161 -72, 2009 Mar � Inducible gene expression: diverse regulatory mechanisms, Nature reviews. Genetics , 2010 Apr 27 � Genome-wide prediction of transcription factor binding sites using an integrated model, Genome biology 11 (1): R 7, 2010 Jan 22 � http: //en. wikipedia. org/wiki/Histone

Q&A
 Nucleosome remodeling
Nucleosome remodeling Nucleosomes
Nucleosomes Positive identification example
Positive identification example Hci patterns may or may not include code for implementation
Hci patterns may or may not include code for implementation Common factors of 18 and 27
Common factors of 18 and 27 Angular frequency to frequency
Angular frequency to frequency Situation producing questions in research examples
Situation producing questions in research examples Lesson 8-2 factoring by gcf
Lesson 8-2 factoring by gcf What is factored form
What is factored form Factor out the greatest common factor
Factor out the greatest common factor Eukaryotic vs prokaryotic transcription
Eukaryotic vs prokaryotic transcription Replikasi dna
Replikasi dna Jml transcription
Jml transcription Termination of transcription in prokaryotes
Termination of transcription in prokaryotes Transcription
Transcription Transcription and translation
Transcription and translation Dna replication transcription and translation
Dna replication transcription and translation Transcription
Transcription Dna transcription and translation
Dna transcription and translation Transcription
Transcription Blood type chart
Blood type chart Dna transcription
Dna transcription Transcription
Transcription Translation
Translation Transcription and translation practice worksheet answer key
Transcription and translation practice worksheet answer key Hiv reverse transcription
Hiv reverse transcription Translation transcription
Translation transcription How to write 115 on a check
How to write 115 on a check Transcription
Transcription Ch
Ch Picture transcription
Picture transcription