Nucleic Acids Metabolism Nitrogenous Bases Planar aromatic and

Nucleic Acids Metabolism
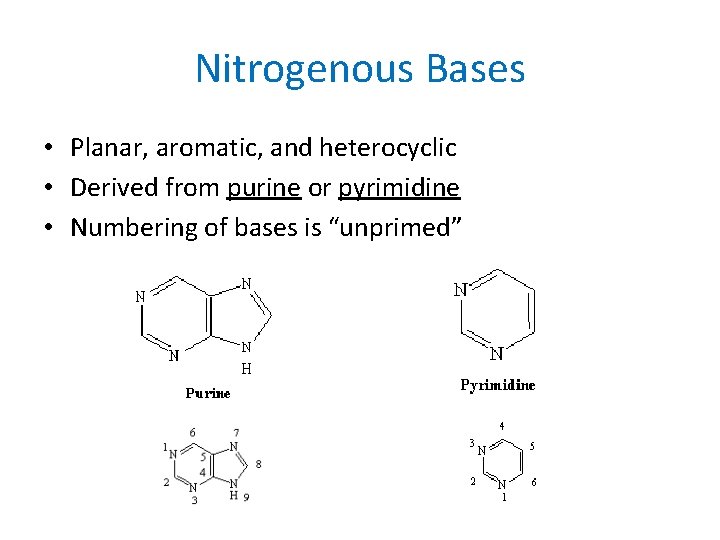
Nitrogenous Bases • Planar, aromatic, and heterocyclic • Derived from purine or pyrimidine • Numbering of bases is “unprimed”
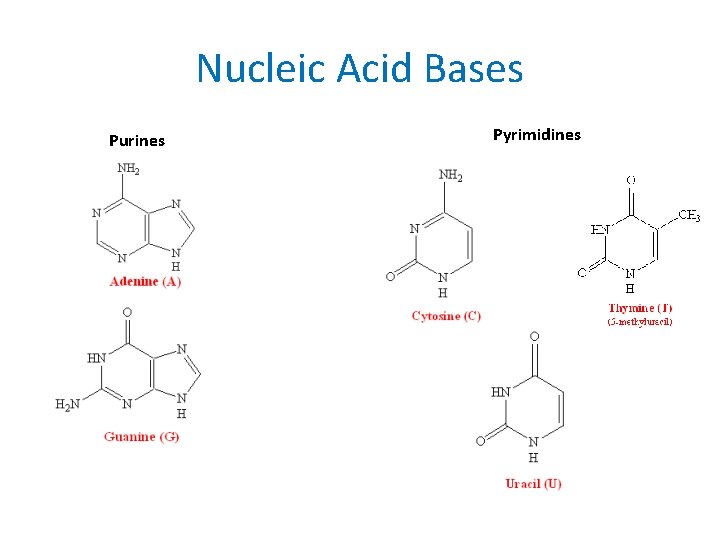
Nucleic Acid Bases Purines Pyrimidines
Sugars • Pentoses (5 -C sugars) • Numbering of sugars is “primed”
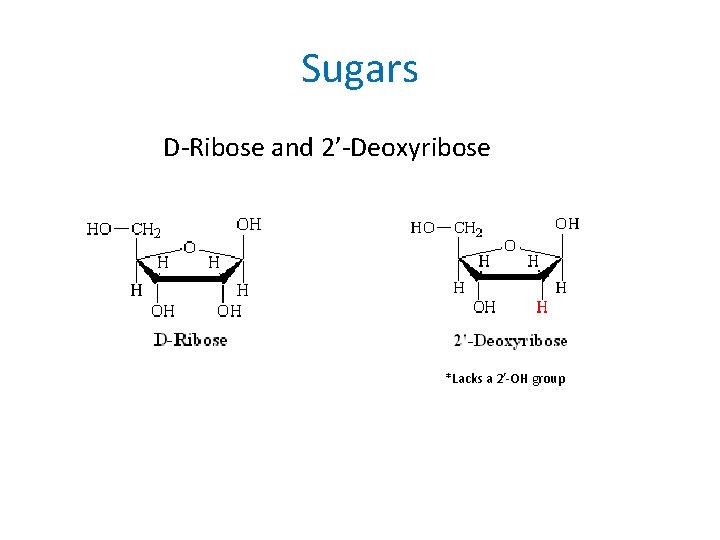
Sugars D-Ribose and 2’-Deoxyribose *Lacks a 2’-OH group
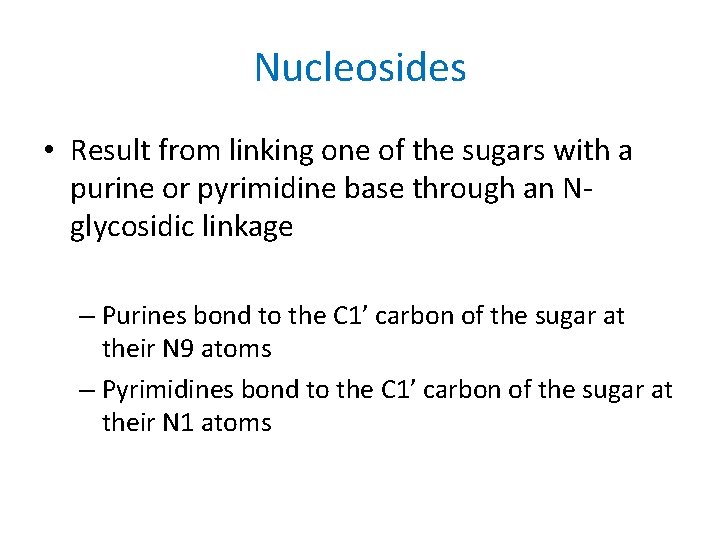
Nucleosides • Result from linking one of the sugars with a purine or pyrimidine base through an Nglycosidic linkage – Purines bond to the C 1’ carbon of the sugar at their N 9 atoms – Pyrimidines bond to the C 1’ carbon of the sugar at their N 1 atoms
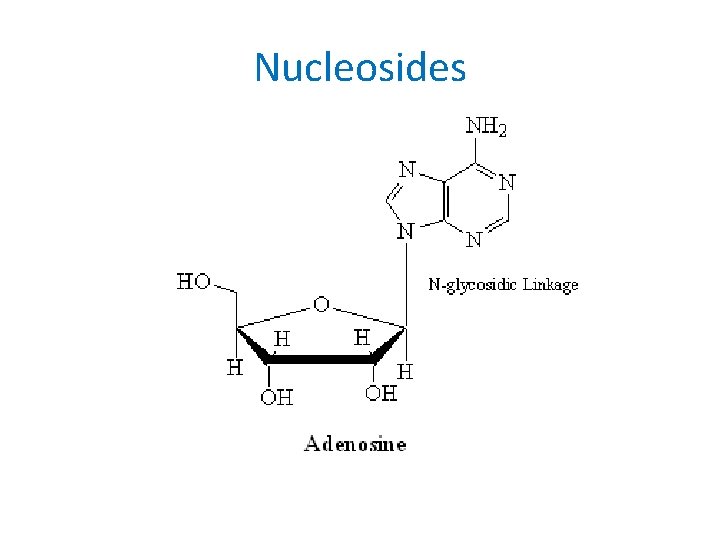
Nucleosides

Phosphate Groups • Mono-, di- or triphosphates • Phosphates can be bonded to either C 3 or C 5 atoms of the sugar
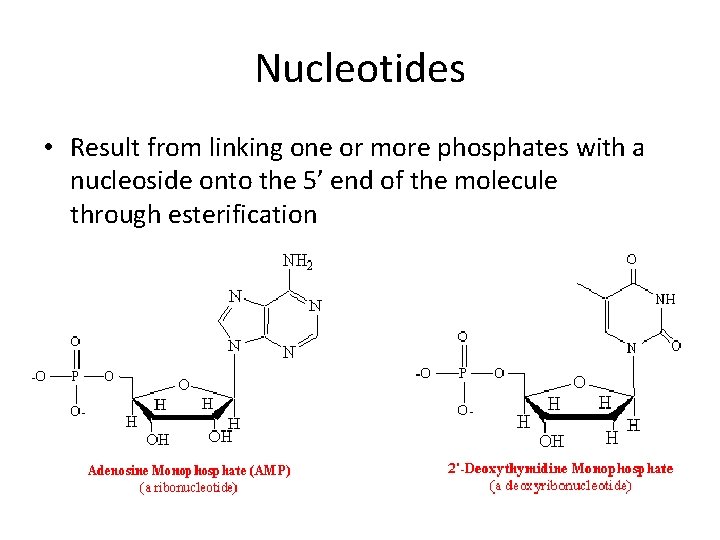
Nucleotides • Result from linking one or more phosphates with a nucleoside onto the 5’ end of the molecule through esterification
Nucleotides • RNA (ribonucleic acid) is a polymer of ribonucleotides • DNA (deoxyribonucleic acid) is a polymer of deoxyribonucleotides • Both deoxy- and ribonucleotides contain Adenine, Guanine and Cytosine – Ribonucleotides contain Uracil – Deoxyribonucleotides contain Thymine
Nucleotides • Monomers for nucleic acid polymers • Nucleoside Triphosphates are important energy carriers (ATP, GTP) • Important components of coenzymes – FAD, NAD+ and Coenzyme A

Naming Conventions • Nucleosides: – Purine nucleosides end in “-sine” • Adenosine, Guanosine – Pyrimidine nucleosides end in “-dine” • Thymidine, Cytidine, Uridine • Nucleotides: – Start with the nucleoside name from above and add “mono-”, “di-”, or “triphosphate” • Adenosine Monophosphate, Cytidine Triphosphate, Deoxythymidine Diphosphate
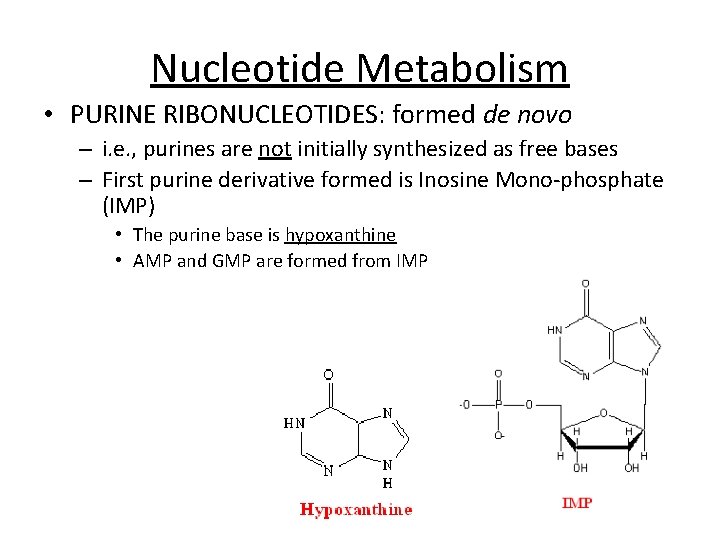
Nucleotide Metabolism • PURINE RIBONUCLEOTIDES: formed de novo – i. e. , purines are not initially synthesized as free bases – First purine derivative formed is Inosine Mono-phosphate (IMP) • The purine base is hypoxanthine • AMP and GMP are formed from IMP

Purine Nucleotides • Get broken down into Uric Acid (a purine) Buchanan (mid 1900 s) showed where purine ring components came from: N 1: Aspartate Amine C 2, C 8: Formate N 3, N 9: Glutamine C 4, C 5, N 7: Glycine C 6: Bicarbonate Ion
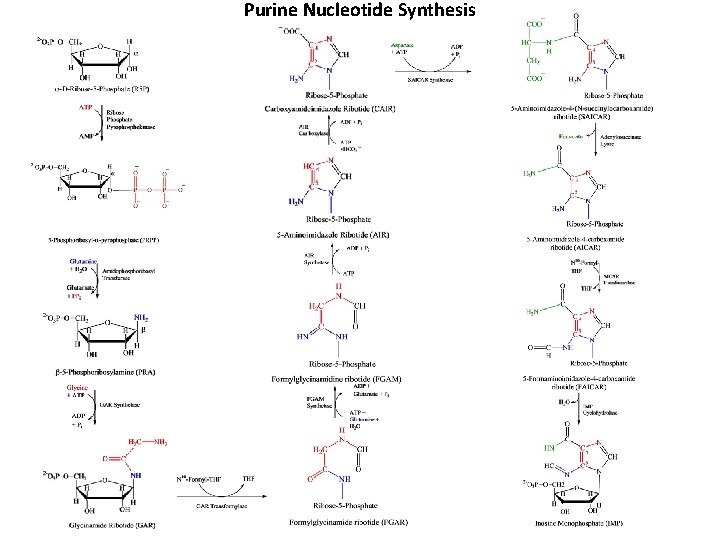
Purine Nucleotide Synthesis
Purine Nucleotide Synthesis at a Glance • ATP is involved in 6 steps • PRPP in the first step of Purine synthesis is also a precursor for Pyrimidine Synthesis, His and Trp synthesis – Role of ATP in first step is unique– group transfer rather than coupling • In second step, C 1 notation changes from a to b (anomers specifying OH positioning on C 1 with respect to C 4 group) • In step 2, PPi is hydrolyzed to 2 Pi (irreversible, “committing” step)
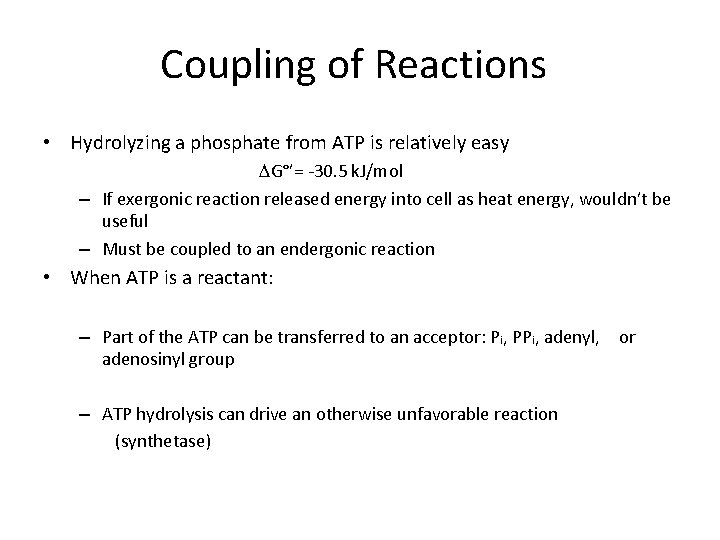
Coupling of Reactions • Hydrolyzing a phosphate from ATP is relatively easy G°’= -30. 5 k. J/mol – If exergonic reaction released energy into cell as heat energy, wouldn’t be useful – Must be coupled to an endergonic reaction • When ATP is a reactant: – Part of the ATP can be transferred to an acceptor: Pi, PPi, adenyl, or adenosinyl group – ATP hydrolysis can drive an otherwise unfavorable reaction (synthetase)

Purine Biosynthetic Pathway • Channeling of some reactions on pathway organizes and controls processing of substrates to products in each step – Increases overall rate of pathway and protects intermediates from degradation • In animals, IMP synthesis pathway shows channeling at: – Reactions 3, 4, 6 – Reactions 7, 8 – Reactions 10, 11

IMP Conversion to AMP
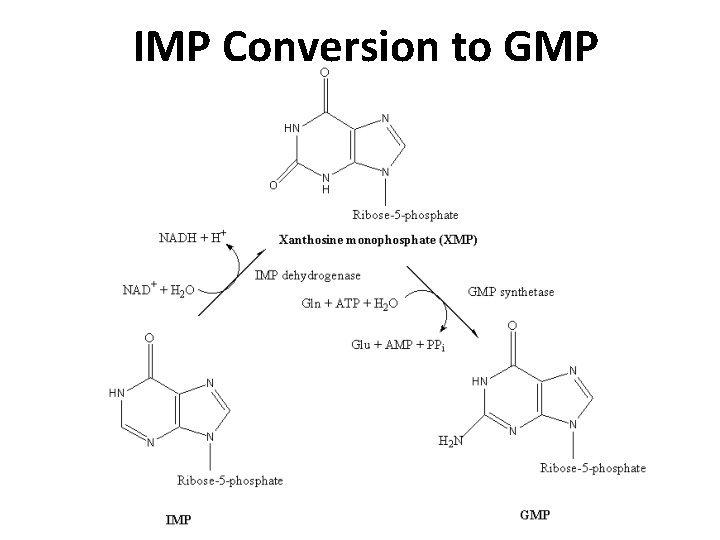
IMP Conversion to GMP
Regulatory Control of Purine Nucleotide Biosynthesis • GTP is involved in AMP synthesis and ATP is involved in GMP synthesis (reciprocal control of production) • PRPP is a biosynthetically “central” molecule (why? ) – ADP/GDP levels – negative feedback on Ribose Phosphate Pyrophosphokinase – Amidophosphoribosyl transferase is activated by PRPP levels – APRT activity has negative feedback at two sites • ATP, ADP, AMP bound at one site • GTP, GDP AND GMP bound at the other site • Rate of AMP production increases with increasing concentrations of GTP; rate of GMP production increases with increasing concentrations of ATP
Regulatory Control of Purine Biosynthesis • Above the level of IMP production: – Independent control – Synergistic control – Feedforward activation by PRPP • Below level of IMP production – Reciprocal control • Total amounts of purine nucleotides controlled • Relative amounts of ATP, GTP controlled

Purine Catabolism and Salvage • All purine degradation leads to uric acid • Ingested nucleic acids are degraded to nucleotides by pancreatic nucleases, and intestinal phosphodiesterases in the intestine • Group-specific nucleotidases and non-specific phosphatases degrade nucleotides into nucleosides – Direct absorption of nucleosides – Further degradation Nucleoside + H 2 O base + ribose (nucleosidase) Nucleoside + Pi base + r-1 -phosphate (n. phosphorylase) NOTE: MOST INGESTED NUCLEIC ACIDS ARE DEGRADED AND EXCRETED.
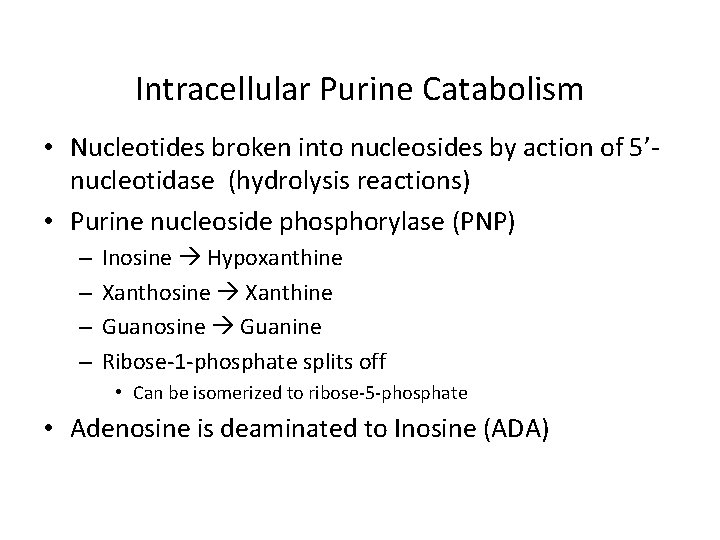
Intracellular Purine Catabolism • Nucleotides broken into nucleosides by action of 5’nucleotidase (hydrolysis reactions) • Purine nucleoside phosphorylase (PNP) – – Inosine Hypoxanthine Xanthosine Xanthine Guanosine Guanine Ribose-1 -phosphate splits off • Can be isomerized to ribose-5 -phosphate • Adenosine is deaminated to Inosine (ADA)
Intracellular Purine Catabolism • Xanthine is the point of convergence for the metabolism of the purine bases • Xanthine Uric acid – Xanthine oxidase catalyzes two reactions • Purine ribonucleotide degradation pathway is same for purine deoxyribonucleotides
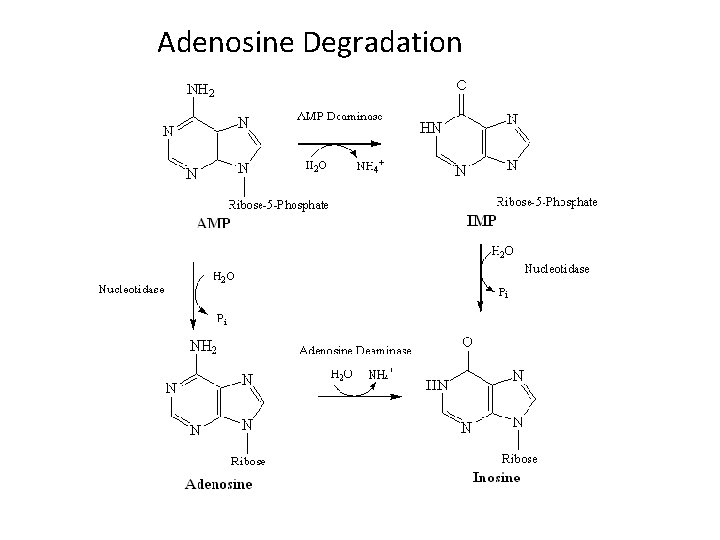
Adenosine Degradation
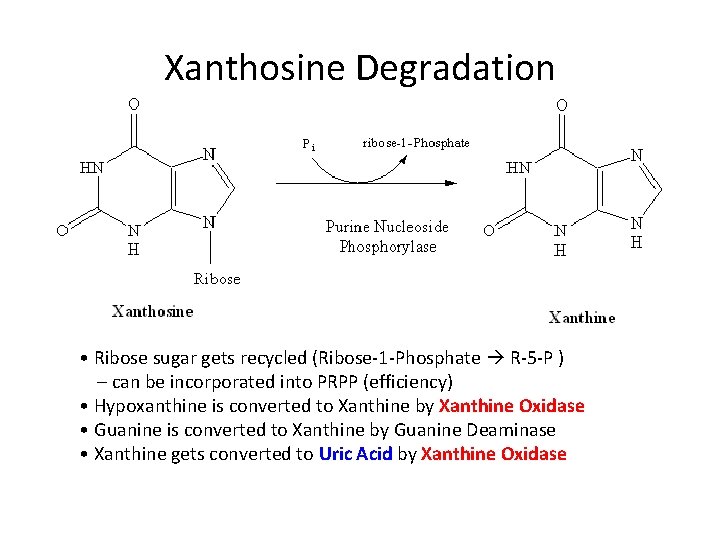
Xanthosine Degradation • Ribose sugar gets recycled (Ribose-1 -Phosphate R-5 -P ) – can be incorporated into PRPP (efficiency) • Hypoxanthine is converted to Xanthine by Xanthine Oxidase • Guanine is converted to Xanthine by Guanine Deaminase • Xanthine gets converted to Uric Acid by Xanthine Oxidase
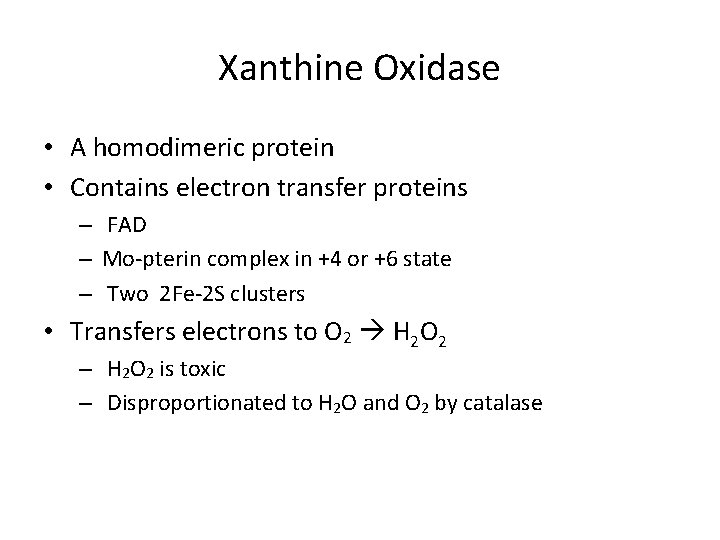
Xanthine Oxidase • A homodimeric protein • Contains electron transfer proteins – FAD – Mo-pterin complex in +4 or +6 state – Two 2 Fe-2 S clusters • Transfers electrons to O 2 H 2 O 2 – H 2 O 2 is toxic – Disproportionated to H 2 O and O 2 by catalase
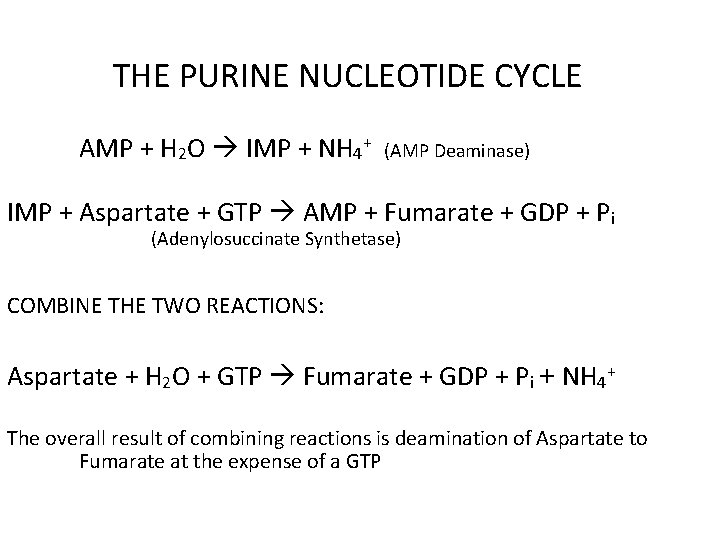
THE PURINE NUCLEOTIDE CYCLE AMP + H 2 O IMP + NH 4+ (AMP Deaminase) IMP + Aspartate + GTP AMP + Fumarate + GDP + Pi (Adenylosuccinate Synthetase) COMBINE THE TWO REACTIONS: Aspartate + H 2 O + GTP Fumarate + GDP + Pi + NH 4+ The overall result of combining reactions is deamination of Aspartate to Fumarate at the expense of a GTP
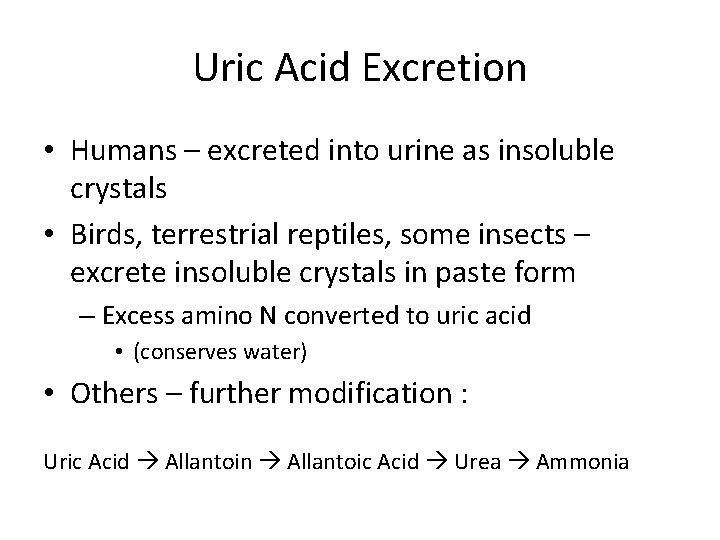
Uric Acid Excretion • Humans – excreted into urine as insoluble crystals • Birds, terrestrial reptiles, some insects – excrete insoluble crystals in paste form – Excess amino N converted to uric acid • (conserves water) • Others – further modification : Uric Acid Allantoin Allantoic Acid Urea Ammonia
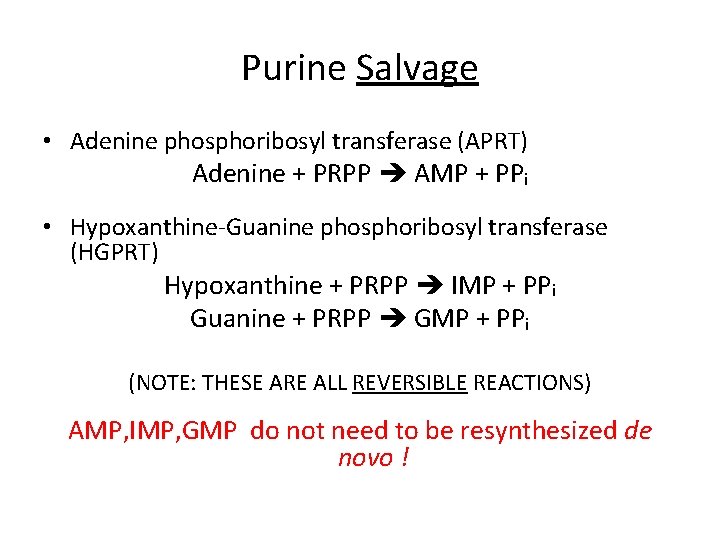
Purine Salvage • Adenine phosphoribosyl transferase (APRT) Adenine + PRPP AMP + PPi • Hypoxanthine-Guanine phosphoribosyl transferase (HGPRT) Hypoxanthine + PRPP IMP + PPi Guanine + PRPP GMP + PPi (NOTE: THESE ARE ALL REVERSIBLE REACTIONS) AMP, IMP, GMP do not need to be resynthesized de novo !
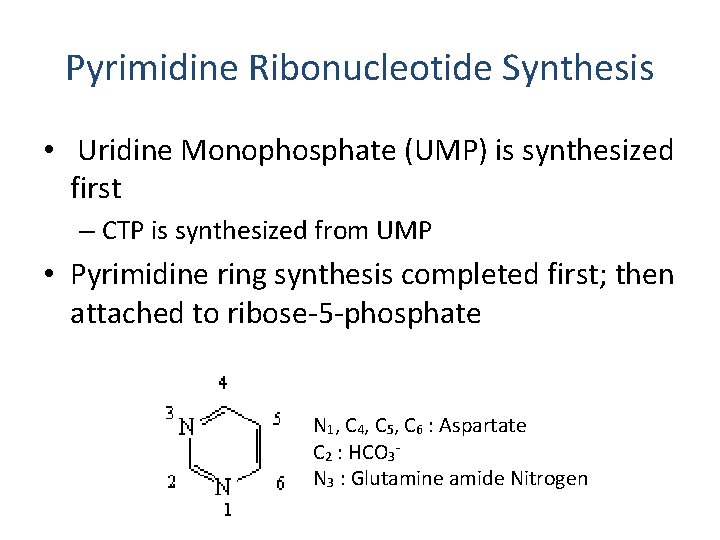
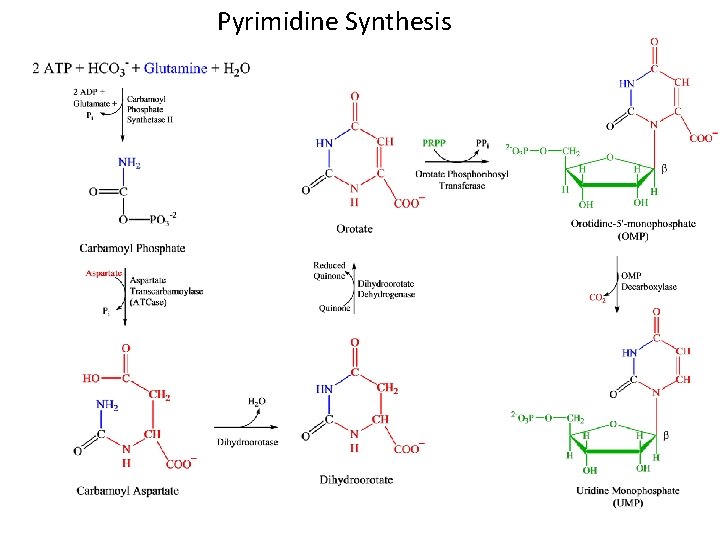
Pyrimidine Ribonucleotide Synthesis • Uridine Monophosphate (UMP) is synthesized first – CTP is synthesized from UMP • Pyrimidine ring synthesis completed first; then attached to ribose-5 -phosphate N 1, C 4, C 5, C 6 : Aspartate C 2 : HCO 3 N 3 : Glutamine amide Nitrogen
Pyrimidine Synthesis

UMP Synthesis Overview • 2 ATPs needed: both used in first step – One transfers phosphate, the other is hydrolyzed to ADP and Pi • 2 condensation rxns: form carbamoyl aspartate and dihydroorotate (intramolecular) • Dihydroorotate dehydrogenase is an intra-mitochondrial enzyme; oxidizing power comes from quinone reduction • Attachment of base to ribose ring is catalyzed by OPRT; PRPP provides ribose-5 -P – PPi splits off PRPP – irreversible

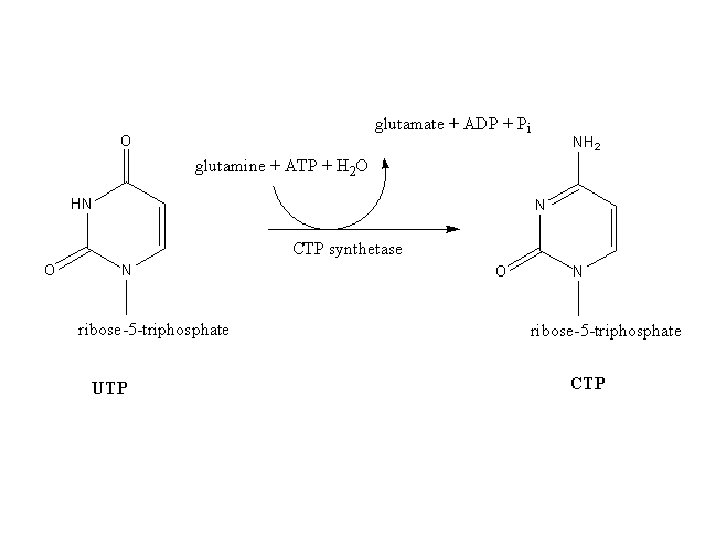
UMP UTP and CTP • Nucleoside monophosphate kinase catalyzes transfer of Pi to UMP to form UDP; nucleoside diphosphate kinase catalyzes transfer of Pi from ATP to UDP to form UTP • CTP formed from UTP via CTP Synthetase driven by ATP hydrolysis – Glutamine provides amide nitrogen for C 4 in animals
Regulatory Control of Pyrimidine Synthesis • Differs between bacteria and animals – Bacteria – regulation at ATCase rxn • Animals – regulation at carbamoyl phosphate synthetase II – UDP and UTP inhibit enzyme; ATP and PRPP activate it – UMP and CMP competitively inhibit OMP Decarboxylase *Purine synthesis inhibited by ADP and GDP at ribose phosphate pyrophosphokinase step, controlling level of PRPP also regulates pyrimidines
Degradation of Pyrimidines • CMP and UMP degraded to bases similarly to purines – Dephosphorylation – Deamination – Glycosidic bond cleavage • Uracil reduced in liver, forming b-alanine – Converted to malonyl-Co. A fatty acid synthesis for energy metabolism
Deoxyribonucleotide Formation • Purine/Pyrimidine degradation are the same for ribonucleotides and deoxyribonucleotides • Biosynthetic pathways are only for ribonucleotide production • Deoxyribonucleotides are synthesized from corresponding ribonucleotides
DNA vs. RNA: REVIEW • DNA composed of deoxyribonucleotides • Ribose sugar in DNA lacks hydroxyl group at 2’ Carbon • Uracil doesn’t (normally) appear in DNA – Thymine (5 -methyluracil) appears instead
Formation of Deoxyribonucleotides • Reduction of 2’ carbon done via a free radical mechanism catalyzed by “Ribonucleotide Reductases” – E. coli RNR reduces ribonucleoside diphosphates (NDPs) to deoxyribonucleoside diphosphates (d. NDPs) • Two subunits: R 1 and R 2 – A Heterotetramer: (R 1)2 and (R 2)2
Thymine Formation • Formed by methylating deoxyuridine monophosphate (d. UMP) • UTP is needed for RNA production, but d. UTP not needed for DNA – If d. UTP produced excessively, would cause substitution errors (d. UTP for d. TTP) • d. UTP hydrolyzed by d. UTPase (d. UTP diphosphohydrolase) to d. UMP methylated at C 5 to form d. TMP rephosphorylate to form d. TTP
- Slides: 42