Nucleic Acids and Protein Synthesis Nucleic Acids DNA


















- Slides: 46

Nucleic Acids and Protein Synthesis

Nucleic Acids • DNA • Deoxyribonucleic Acid • RNA • Ribonucleic Acid

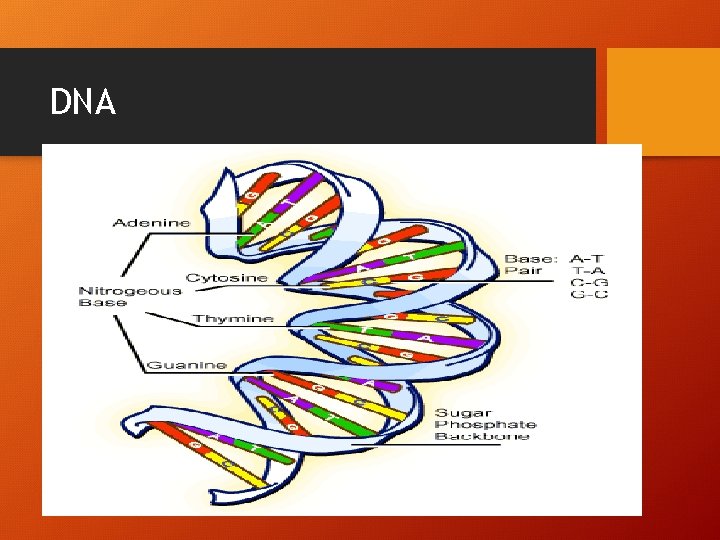
DNA • Double stranded helix • Never leaves the nucleus • Watson, Crick, Wilkins won Nobel Prize in 1962 • Franklin died in 1958 never recognized

DNA

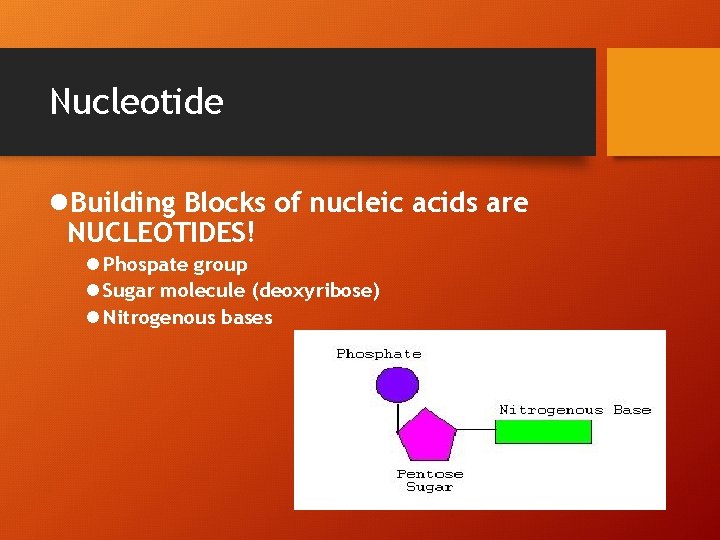
Nucleotide l. Building Blocks of nucleic acids are NUCLEOTIDES! l Phospate group l Sugar molecule (deoxyribose) l Nitrogenous bases

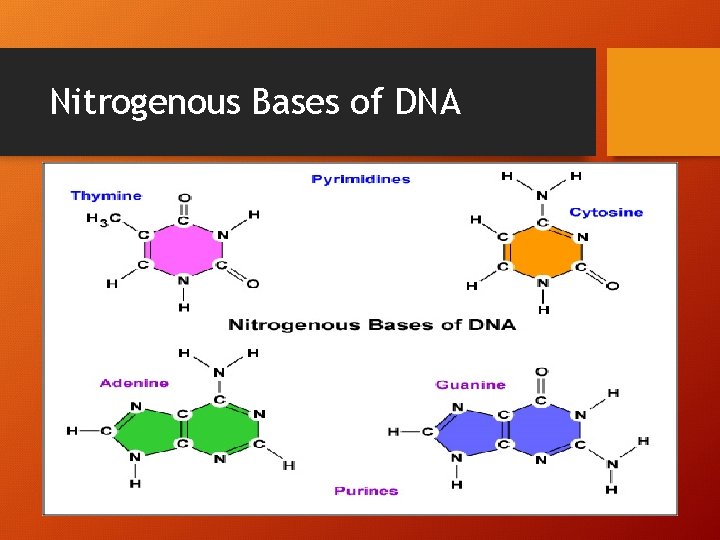
Nitrogenous Bases of DNA

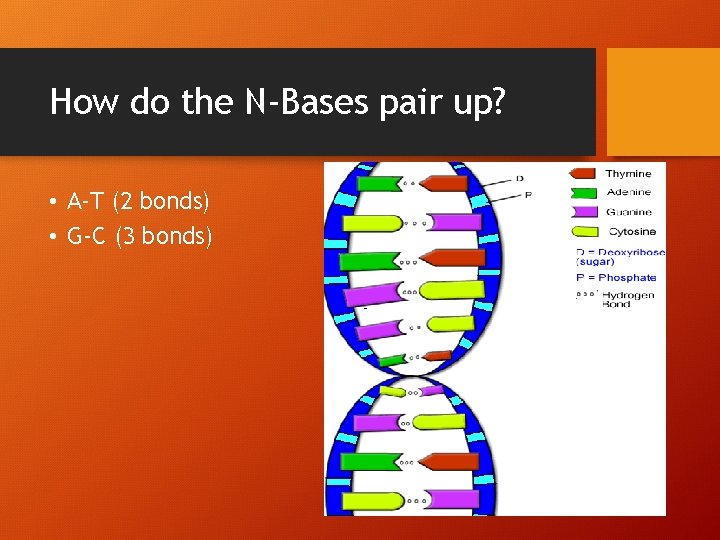
How do the N-Bases pair up? • A-T (2 bonds) • G-C (3 bonds)

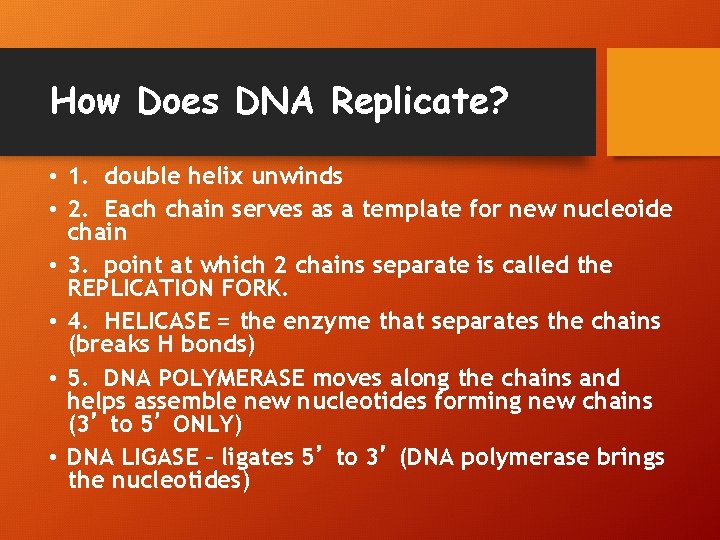
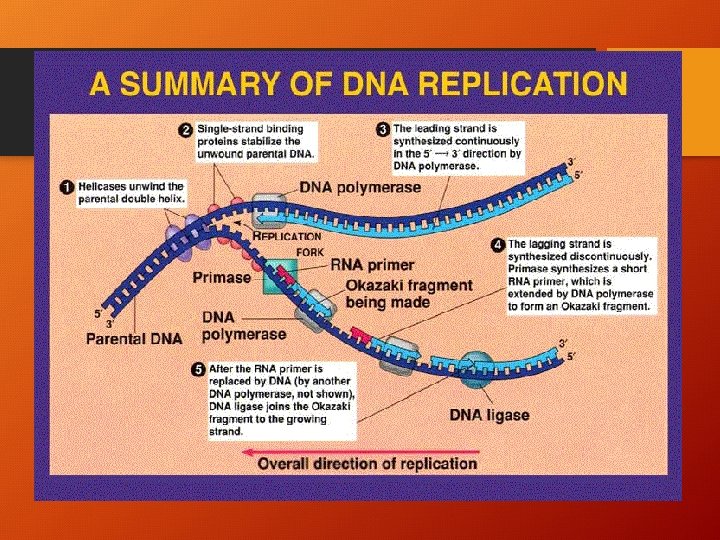
How Does DNA Replicate? • 1. double helix unwinds • 2. Each chain serves as a template for new nucleoide chain • 3. point at which 2 chains separate is called the REPLICATION FORK. • 4. HELICASE = the enzyme that separates the chains (breaks H bonds) • 5. DNA POLYMERASE moves along the chains and helps assemble new nucleotides forming new chains (3’ to 5’ ONLY) • DNA LIGASE – ligates 5’ to 3’ (DNA polymerase brings the nucleotides)

DNA replication continue… • The 3’ sugar has an –OH GROUP • The 5’ sugar has a PHOSPHATE GROUP • LEADING STRAND – formed from 3’-5’ • LAGGING STRAND – formed from 5’- 3’ with the help of DNA LIGASE! • OKAZAKI FRAGMENTS – fragments that will be ligated together

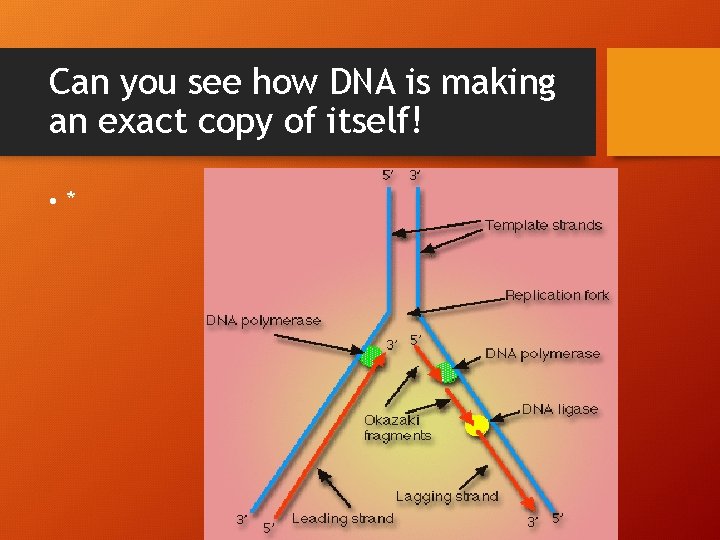
Can you see how DNA is making an exact copy of itself! • *

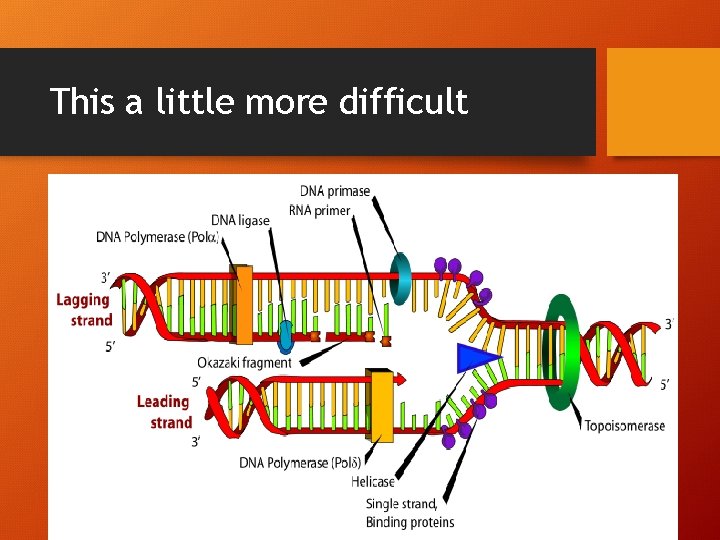
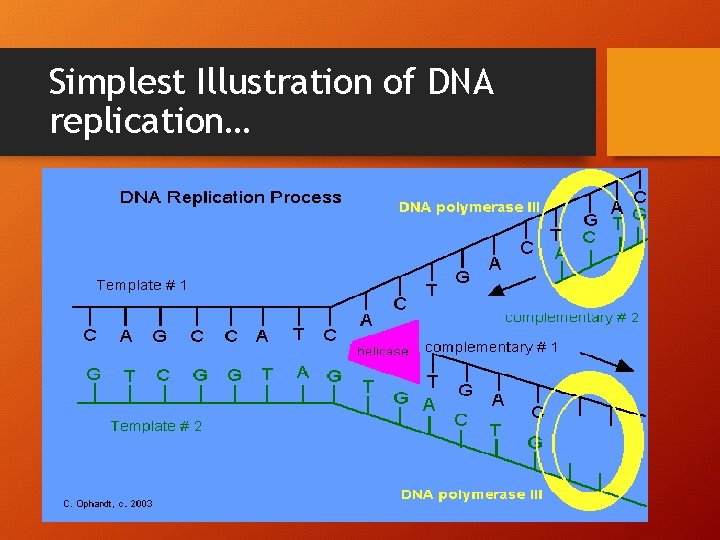
This a little more difficult • Can you figure out the diagram?


Simplest Illustration of DNA replication…

What is a • mutation? ? A CHANGE in the nucleotide sequence at even ONE location!!

Protection l. About 1 in a billion nucleotides in DNA is INCORRECTLY paired. l. DNA polymerase proofreads and removes nucleotides that base pair incorrectly. l. DNA polymerase & DNA ligase also repair damage caused by ultraviolet light, xrays, and toxic chemicals

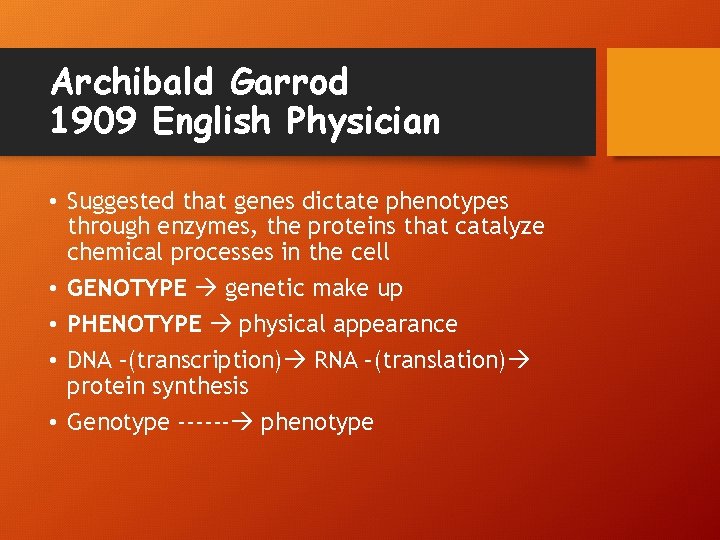
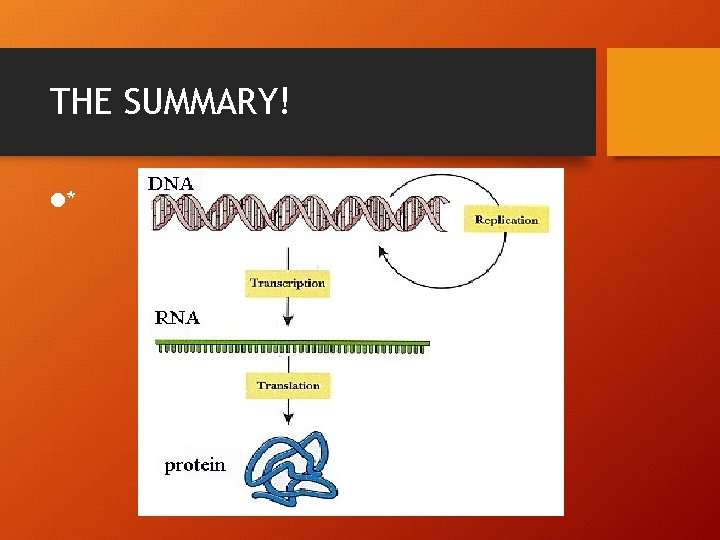
Archibald Garrod 1909 English Physician • Suggested that genes dictate phenotypes through enzymes, the proteins that catalyze chemical processes in the cell • GENOTYPE genetic make up • PHENOTYPE physical appearance • DNA –(transcription) RNA –(translation) protein synthesis • Genotype ------ phenotype

Garrod 1900’s • Children -> defect in 2 a. a. due to defect in the enz. That helps make the a. a • Phenylalanine->PKU • Tyrosine ->albinism • Gene enzyme amino acid (can’t be made) • Couldn’t prove it due to lack of technology

George Beadle & Edward Tatum 1940’s American Geneticists l. ONE GENE ONE ENZYME (polypeptide) HYPOTHESIS: the function of a gene is to dictate the production of a specific enzyme. l. Experimented with bread mold lacked an enzyme in a metabolic pathway that produced some molecules that mold needed to produce an amino acid called arginine.

Tatum & Beadle 1958 Nobel Prize • Proved Garrod correct • Bread mold -> can make all of it’s own a. a. that it needs • Gene -> enzyme -> amino acid • One gene = enzyme • One gene = one protein!!

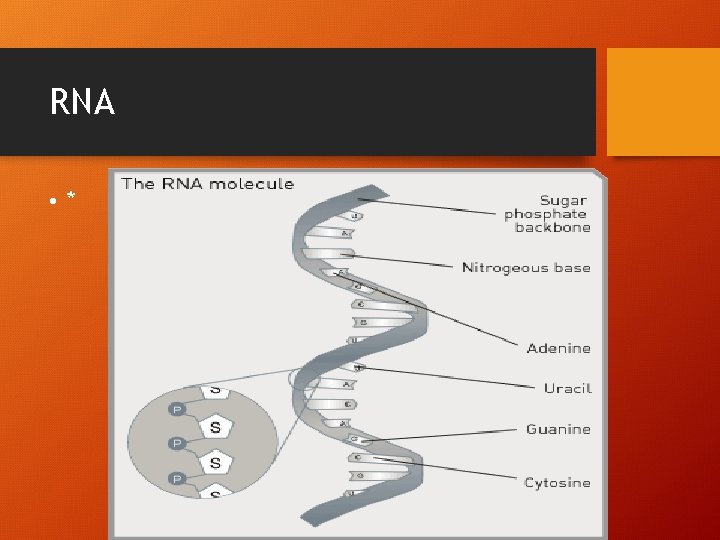
RNA RIBONUCLEIC ACID • SINGLE STRANDED • RESPONSIBLE FOR BRINGING THE GENETIC INFO. FROM THE NUCLEUS TO THE CYTOSOL!

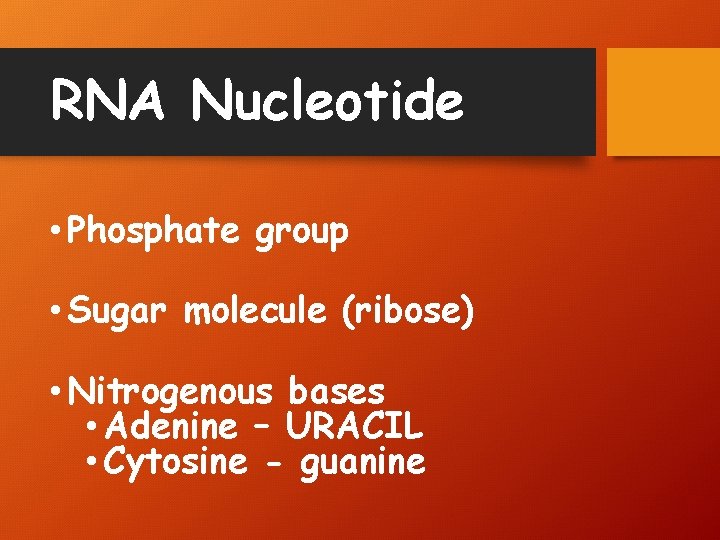
RNA Nucleotide • Phosphate group • Sugar molecule (ribose) • Nitrogenous bases • Adenine – URACIL • Cytosine - guanine

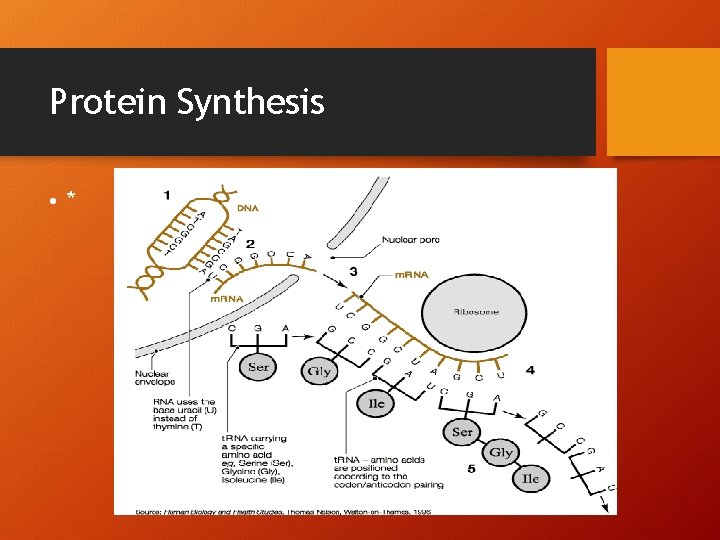
3 Kinds of RNA • m. RNA – (messenger) brings info from DNA in nucleus to cytosol in eukaryotic cells (uncoiled) • t. RNA –(transfer) brings amino acids to m. RNA for translation (hairpin shape) • r. RNA –(ribosomal) most abundant, r. RNA makes up the ribosomes where proteins are made (globular)

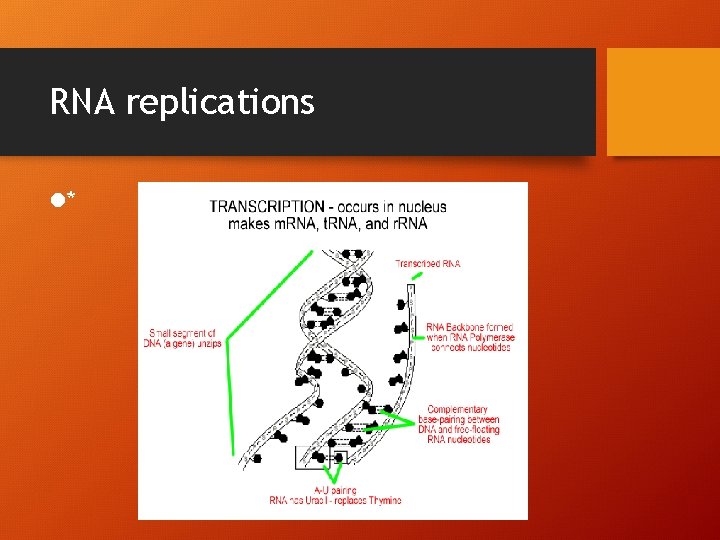
TRANSCRIPTION!! DNA RNA • 1. RNA polymerase-initiates transcription by binding to region on DNA called PROMOTER (causes DNA to separate)-INITIATION PHASE • 2. only ONE of the DNA chains will be used for transcription it’s called the TEMPLATE (promoter dictates which of the two strands will be used) • 3. RNA POLYMERASE – attached to first DNA nucleotide of template chain – then begins adding complementary RNA nucleotides-ELONGATION PHASE

Cont. Transcription • 4. transcription continues until RNA polymerase reaches a TERMINATION SIGNAL on the DNA-TERMINATION PHASE • 5. RNA polymerase releases both the DNA mol. And newly formed RNA mol. Are transcribed in this way (all three!!!)


RNA replications l*

PROKARYOTES • Transcription and translation occur in the SAME place! • NO NUCLEUS!

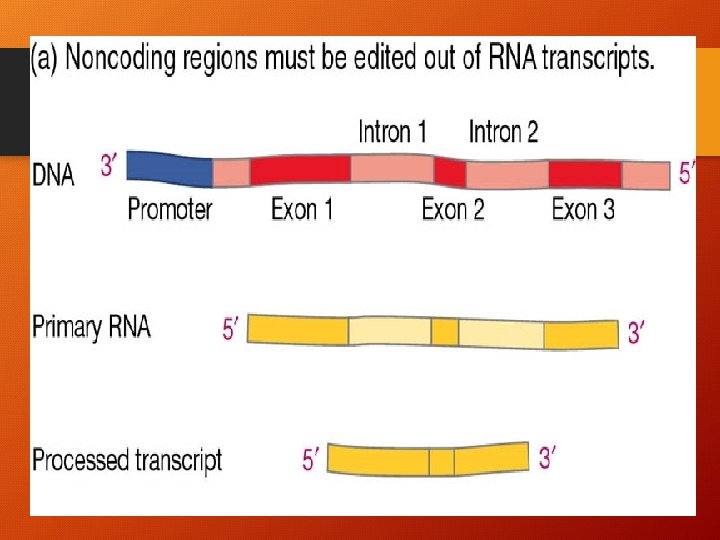
Eukaryotes • 1. Before RNA leaves the nucleus: • G (guanine) cap is attached • A (adenine) tail is attached “many” • 2. These protect the RNA from attack by cellular enzymes and help ribosomes to recognize the m. RNA (cap & tail are NOT translated)

Cont. Eukaryotes • 3. INTRONS (non coding sequence) are removed • 4. EXONS (part of gene that are expressed) are joined to produce a m. RNA molecule with a continuous coding sequence. • NOW RNA CAN LEAVE THE NUCLEUS!


Protein Synthesis • PROTEINS CARRY OUT THE GENETIC INSTRUCTIONS ENCODED IN AN ORGANISM’S DNA!!!!

TRANSLATION The process of assembling from info. Encoded in a m. RNA! • 1. m. RNA leaves nucleus • 2. m. RNA migrates to ribosome in cytosol for protein synthesis • 3. amino acids floating in cytosol are transported to ribosomes by t. RNA molecule • 4. peptide bonds join the amino acids to make polypeptide chain

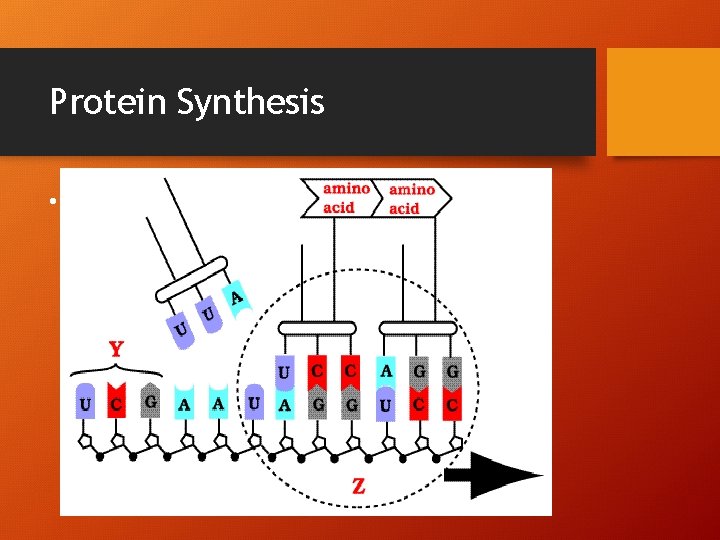
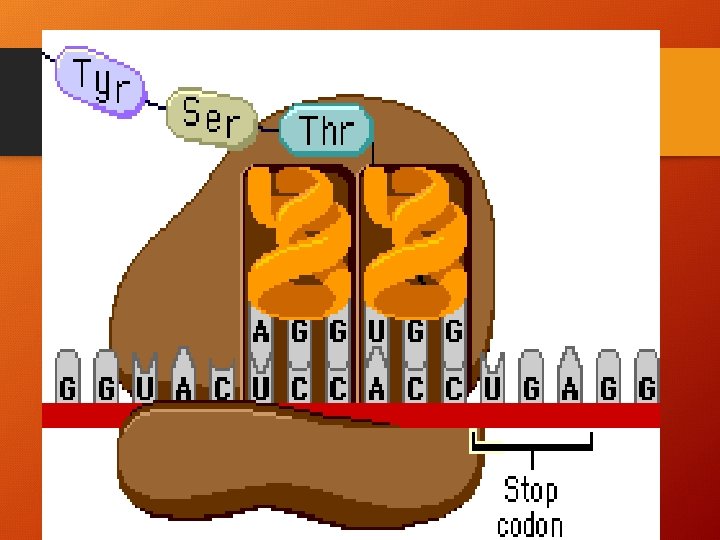
Vocabulary! • 1. GENETIC CODE: correlation between a nucleotide sequence and an amino acid sequence • 2. CODON 3 m. RNA nucleotides, codes for a specific amino acid (64) • 3. START CODON (AUG) & a. a. methionine • 4. STOP CODON (UAA, UAG, UGA) • 5. ANTICODON – 3 t. RNA nucleotides carrying a specific amino acid!

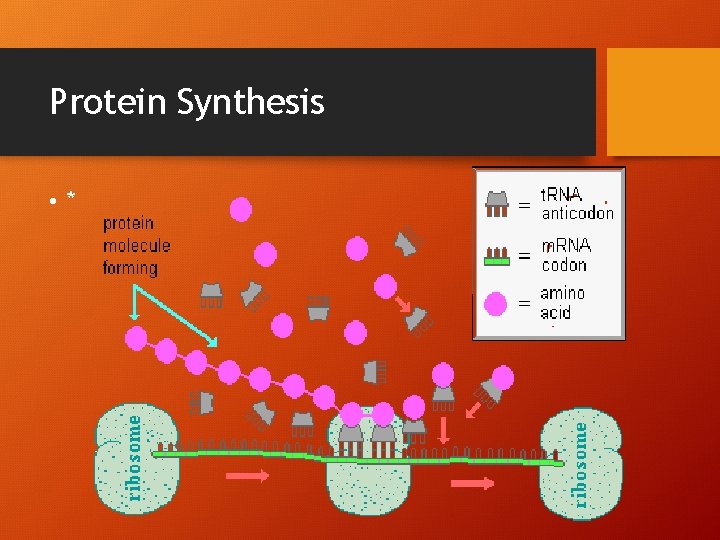
Protein Synthesis • *

Protein Synthesis • *

Protein Synthesis • !

THE SUMMARY! l*

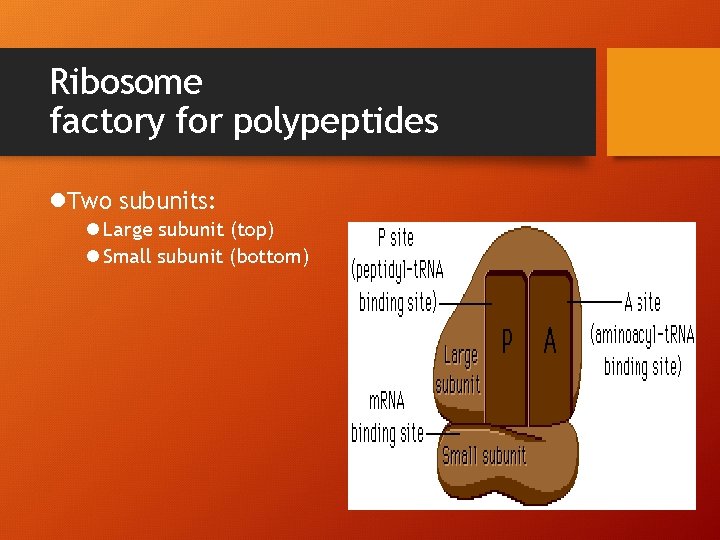
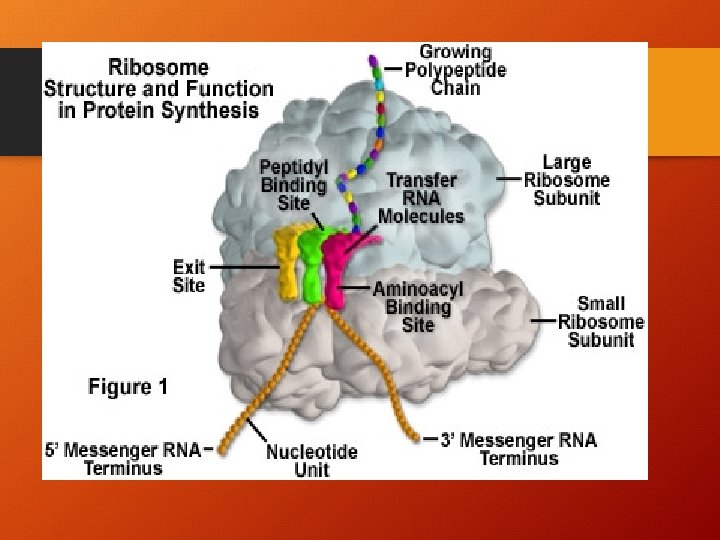
Ribosome factory for polypeptides l. Two subunits: l Large subunit (top) l Small subunit (bottom)

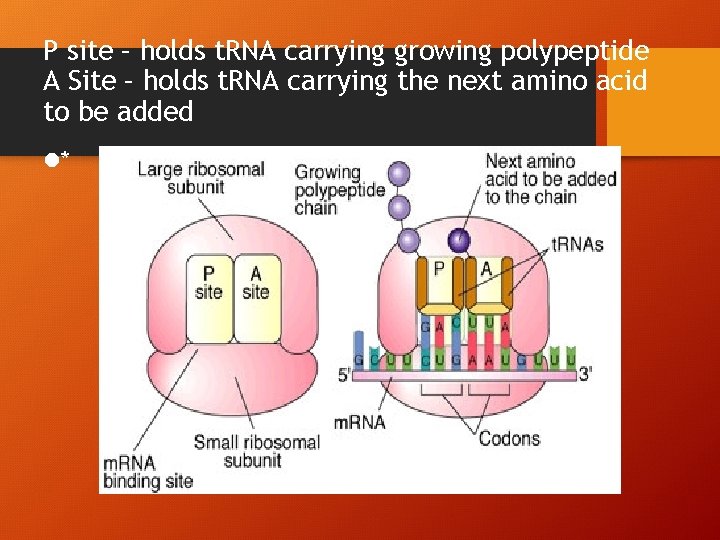
P site – holds t. RNA carrying growing polypeptide A Site – holds t. RNA carrying the next amino acid to be added l*

Initiation Codon Marks the Start of an m. RNA message • 3 PHASES: • 1. INITIATION • 2. ELONGATION • 3. TERMINATION

INITIATION ( 2 steps) l. A) An m. RNA mol. Binds to a small ribosomal subunit. A special initiator t. RNA binds to the specific codon called the START CODON (UAC binds to start codon AUG methionine) l. B) A large ribosomal subunit binds to the small one creating a functional ribosome. The initiator t. RNA fits into the P site of the ribosome.

Elongation and Termination l. Elongation adds amino acids to the polypeptide chain until a stop codon terminates translation.

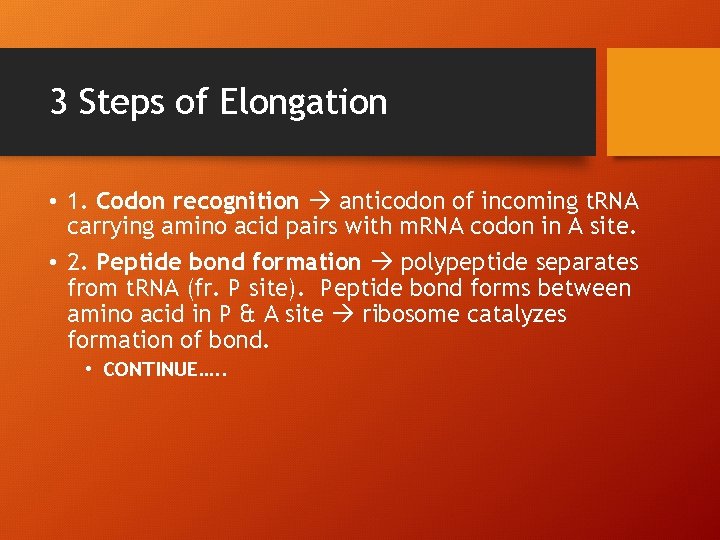
3 Steps of Elongation • 1. Codon recognition anticodon of incoming t. RNA carrying amino acid pairs with m. RNA codon in A site. • 2. Peptide bond formation polypeptide separates from t. RNA (fr. P site). Peptide bond forms between amino acid in P & A site ribosome catalyzes formation of bond. • CONTINUE…. .

CONTINUE… • 3. Translation P site t. RNA now leaves ribosome, the ribosome translocates (moves) the t. RNA in the A site, with its attached polypeptide to the P site. The codon and anticodon remain bonded and the m. RNA and t. RNA move as a unit. This movement brings into the A site the next m. RNA codon to be translated and process can start again! ELONGATION CONTINUES UNTIL A STOP CODON REACHES A SITE.

