Nucleic Acids 28 1 Nucleic Acids Components u




- Slides: 32

Nucleic Acids 28 -1

Nucleic Acids Components u Nucleic acid: A biopolymer containing three types of monomer units. • Heterocyclic aromatic amine bases derived from purine and pyrimidine. • The monosaccharides D-ribose or 2 -deoxy-D-ribose • Phosphoric acid. 28 -2

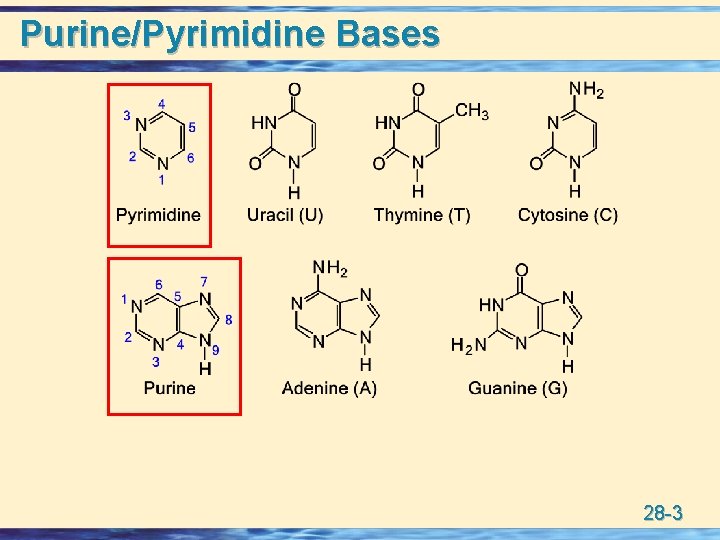
Purine/Pyrimidine Bases 28 -3

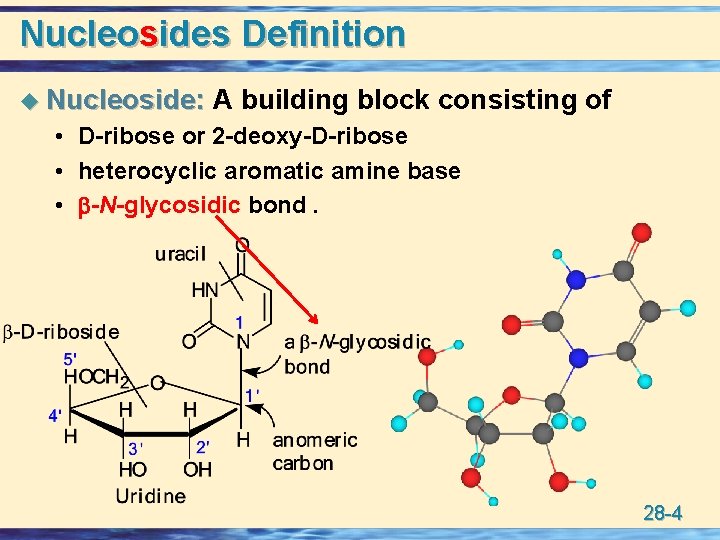
Nucleosides Definition u Nucleoside: A building block consisting of • D-ribose or 2 -deoxy-D-ribose • heterocyclic aromatic amine base • -N-glycosidic bond. 28 -4

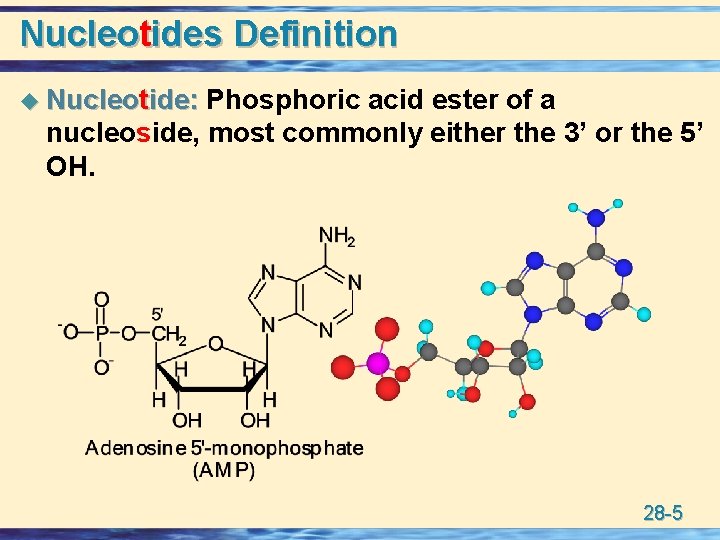
Nucleotides Definition u Nucleotide: Phosphoric acid ester of a nucleoside, most commonly either the 3’ or the 5’ OH. 28 -5

Acyclovir & AZT Used to treat HIV It is used to treat or prevent infections caused by certain kinds of viruses. Examples of these infections include herpes and shingles 28 -7

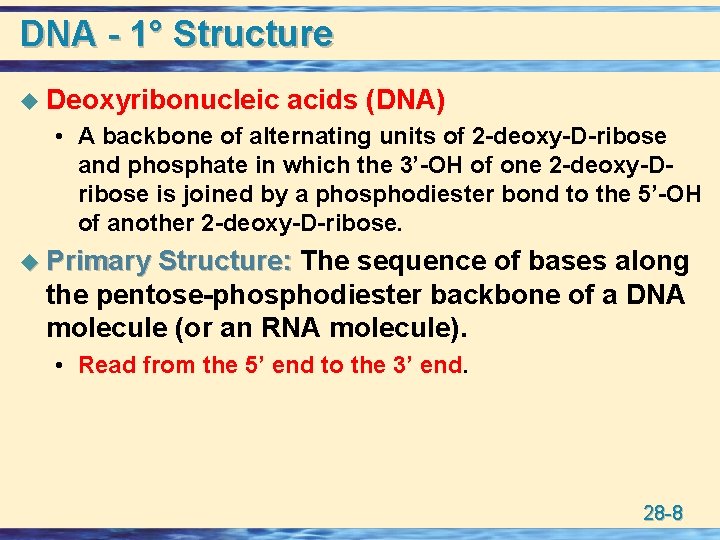
DNA - 1° Structure u Deoxyribonucleic acids (DNA) • A backbone of alternating units of 2 -deoxy-D-ribose and phosphate in which the 3’-OH of one 2 -deoxy-Dribose is joined by a phosphodiester bond to the 5’-OH of another 2 -deoxy-D-ribose. u Primary Structure: The sequence of bases along the pentose-phosphodiester backbone of a DNA molecule (or an RNA molecule). • Read from the 5’ end to the 3’ end. 28 -8

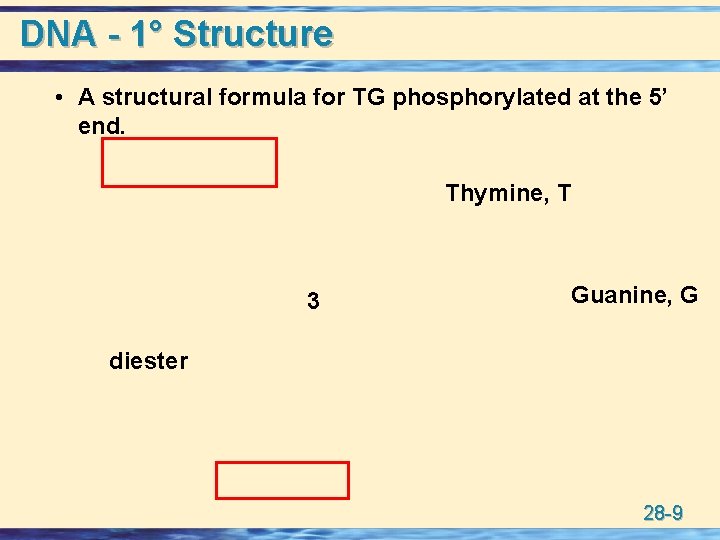
DNA - 1° Structure • A structural formula for TG phosphorylated at the 5’ end. Thymine, T 3 Guanine, G diester 28 -9

DNA - 2° Structure u Secondary structure: The ordered arrangement of nucleic acid strands. u The double helix model of DNA 2° structure was proposed by James Watson and Francis Crick in 1953. u Double helix: A type of 2° structure of DNA molecules in which two antiparallel polynucleotide strands are coiled in a righthanded manner about the same axis. 28 -10

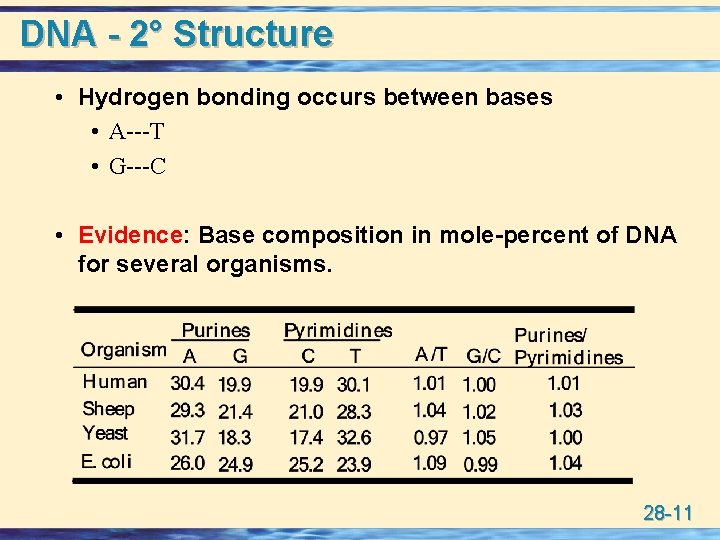
DNA - 2° Structure • Hydrogen bonding occurs between bases • A---T • G---C • Evidence: Base composition in mole-percent of DNA for several organisms. 28 -11

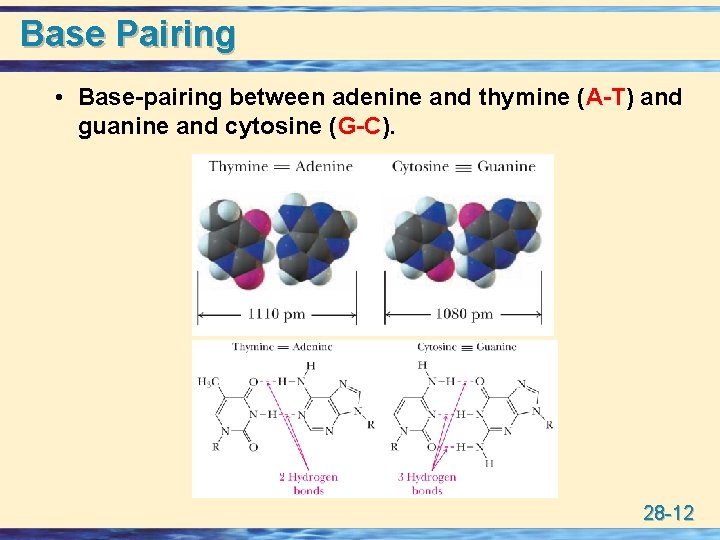
Base Pairing • Base-pairing between adenine and thymine (A-T) and guanine and cytosine (G-C). 28 -12

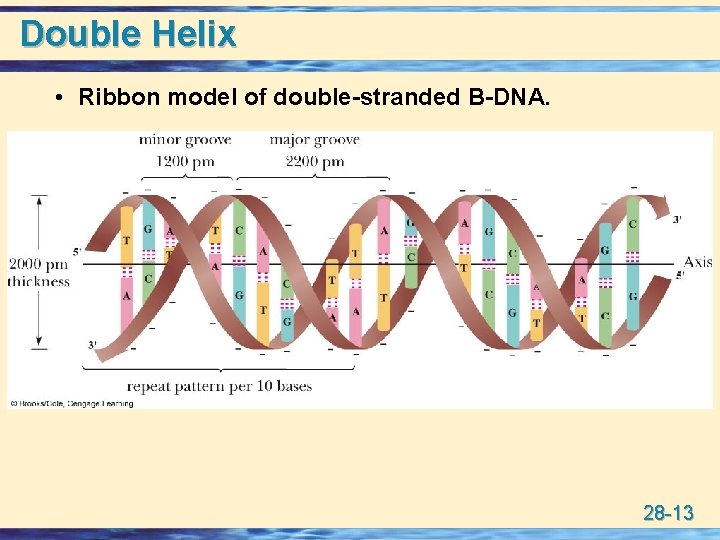
Double Helix • Ribbon model of double-stranded B-DNA. 28 -13

Forms of DNA u B-DNA • • the predominant form in dilute aqueous solution. a right-handed helix. 2000 pm thick with 3400 pm per ten base pairs. minor groove of 1200 pm and major groove of 2200 pm. u A-DNA • a right-handed helix, but thicker than B-DNA. • 2900 pm per 10 base pairs. u Z-DNA • a left-handed double helix. 28 -14

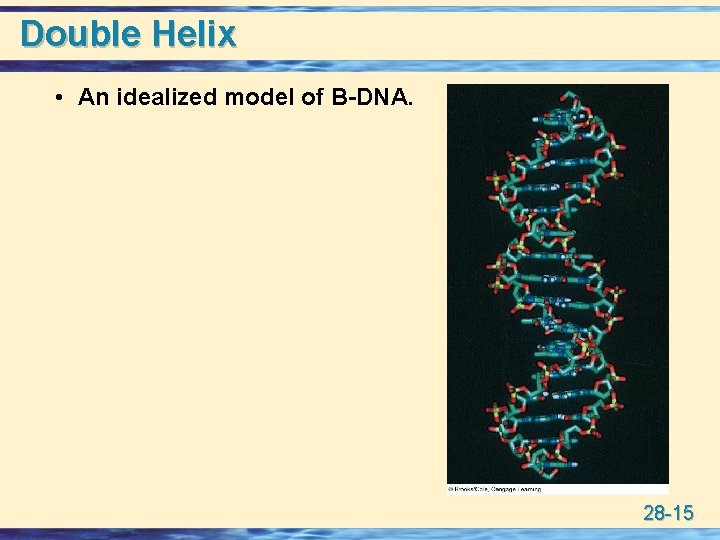
Double Helix • An idealized model of B-DNA. 28 -15

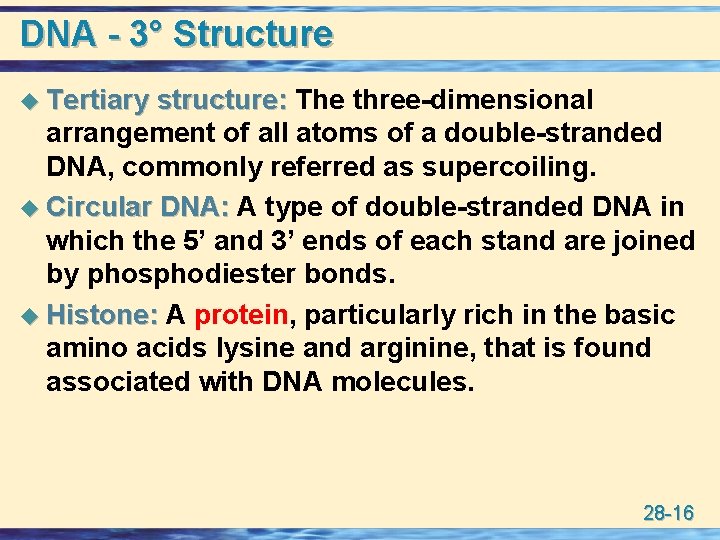
DNA - 3° Structure u Tertiary structure: The three-dimensional arrangement of all atoms of a double-stranded DNA, commonly referred as supercoiling. u Circular DNA: A type of double-stranded DNA in which the 5’ and 3’ ends of each stand are joined by phosphodiester bonds. u Histone: A protein, particularly rich in the basic amino acids lysine and arginine, that is found associated with DNA molecules. 28 -16

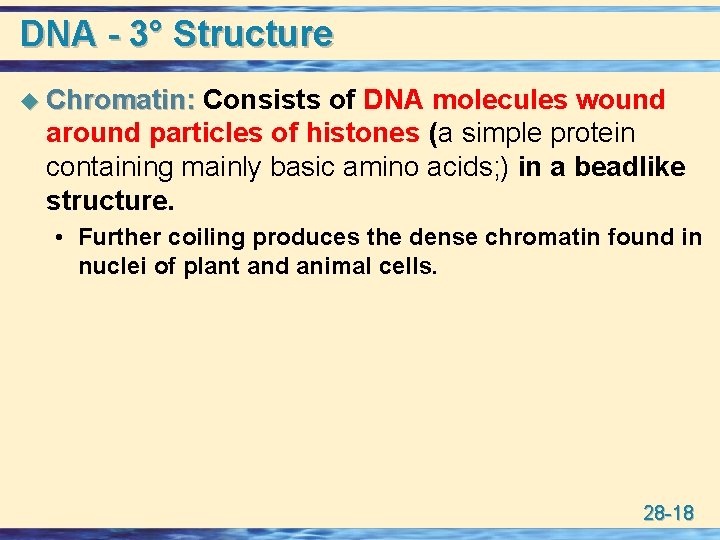
DNA - 3° Structure u Chromatin: Consists of DNA molecules wound around particles of histones (a simple protein containing mainly basic amino acids; ) in a beadlike structure. • Further coiling produces the dense chromatin found in nuclei of plant and animal cells. 28 -18

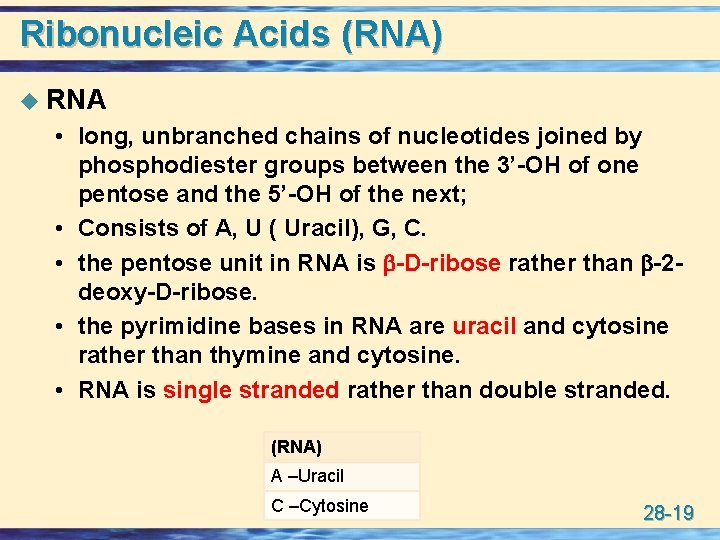
Ribonucleic Acids (RNA) u RNA • long, unbranched chains of nucleotides joined by phosphodiester groups between the 3’-OH of one pentose and the 5’-OH of the next; • Consists of A, U ( Uracil), G, C. • the pentose unit in RNA is -D-ribose rather than -2 deoxy-D-ribose. • the pyrimidine bases in RNA are uracil and cytosine rather than thymine and cytosine. • RNA is single stranded rather than double stranded. (RNA) A –Uracil C –Cytosine 28 -19

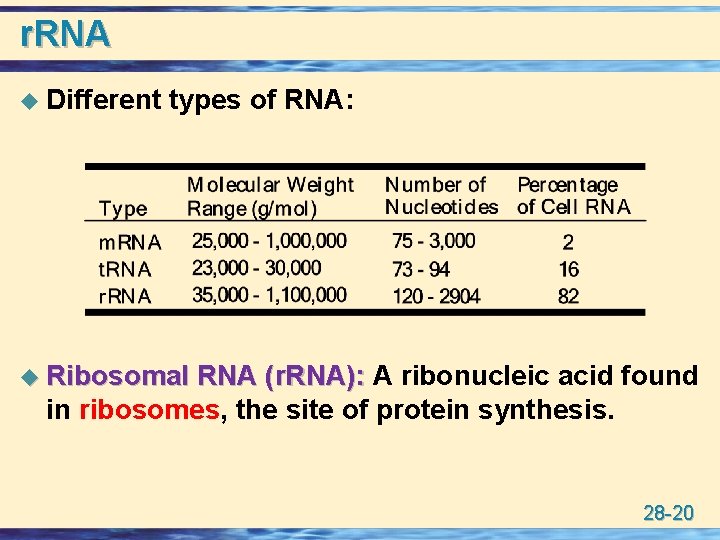
r. RNA u Different types of RNA: u Ribosomal RNA (r. RNA): A ribonucleic acid found in ribosomes, the site of protein synthesis. 28 -20

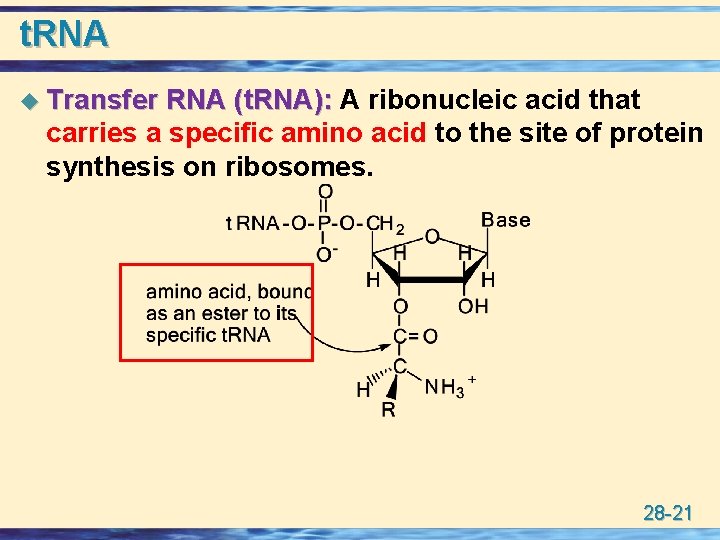
t. RNA u Transfer RNA (t. RNA): A ribonucleic acid that carries a specific amino acid to the site of protein synthesis on ribosomes. 28 -21

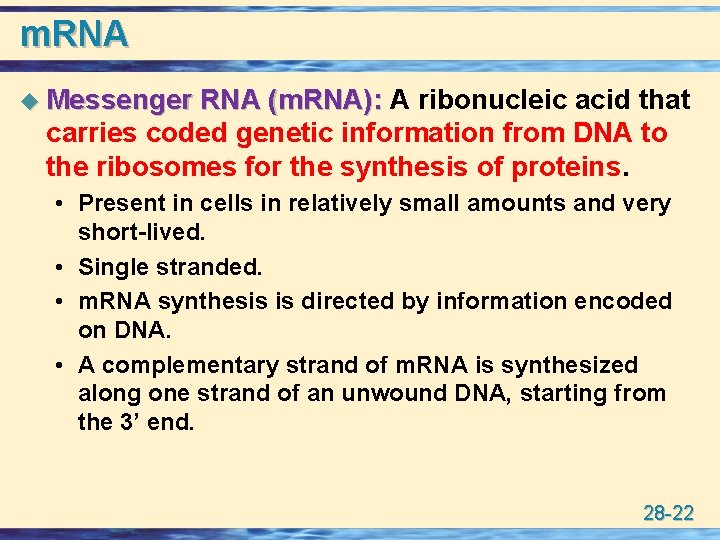
m. RNA u Messenger RNA (m. RNA): A ribonucleic acid that carries coded genetic information from DNA to the ribosomes for the synthesis of proteins. • Present in cells in relatively small amounts and very short-lived. • Single stranded. • m. RNA synthesis is directed by information encoded on DNA. • A complementary strand of m. RNA is synthesized along one strand of an unwound DNA, starting from the 3’ end. 28 -22

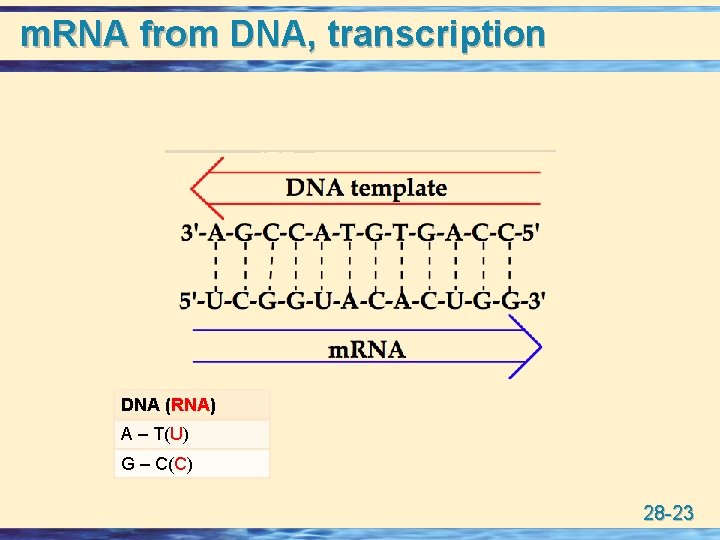
m. RNA from DNA, transcription DNA (RNA) A – T(U) G – C(C) 28 -23

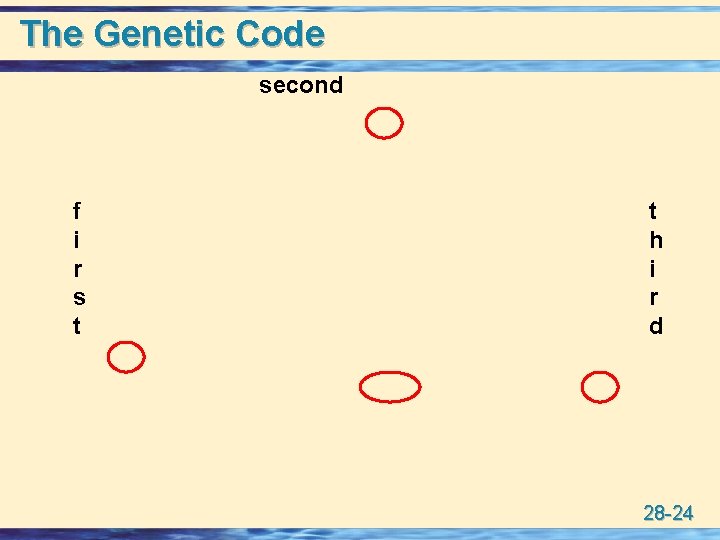
The Genetic Code second f i r s t t h i r d 28 -24

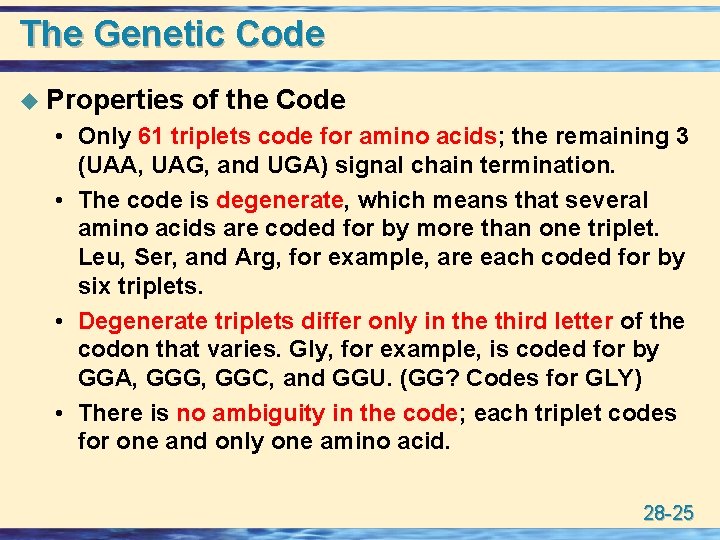
The Genetic Code u Properties of the Code • Only 61 triplets code for amino acids; the remaining 3 (UAA, UAG, and UGA) signal chain termination. • The code is degenerate, which means that several amino acids are coded for by more than one triplet. Leu, Ser, and Arg, for example, are each coded for by six triplets. • Degenerate triplets differ only in the third letter of the codon that varies. Gly, for example, is coded for by GGA, GGG, GGC, and GGU. (GG? Codes for GLY) • There is no ambiguity in the code; each triplet codes for one and only one amino acid. 28 -25

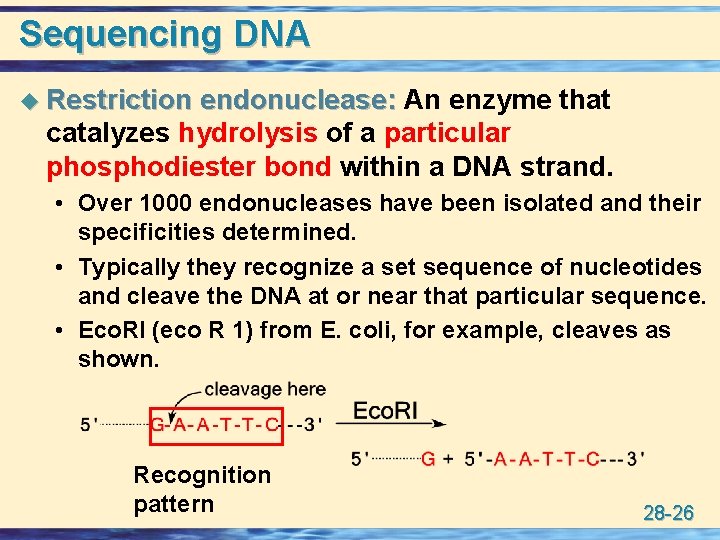
Sequencing DNA u Restriction endonuclease: An enzyme that catalyzes hydrolysis of a particular phosphodiester bond within a DNA strand. • Over 1000 endonucleases have been isolated and their specificities determined. • Typically they recognize a set sequence of nucleotides and cleave the DNA at or near that particular sequence. • Eco. RI (eco R 1) from E. coli, for example, cleaves as shown. Recognition pattern 28 -26

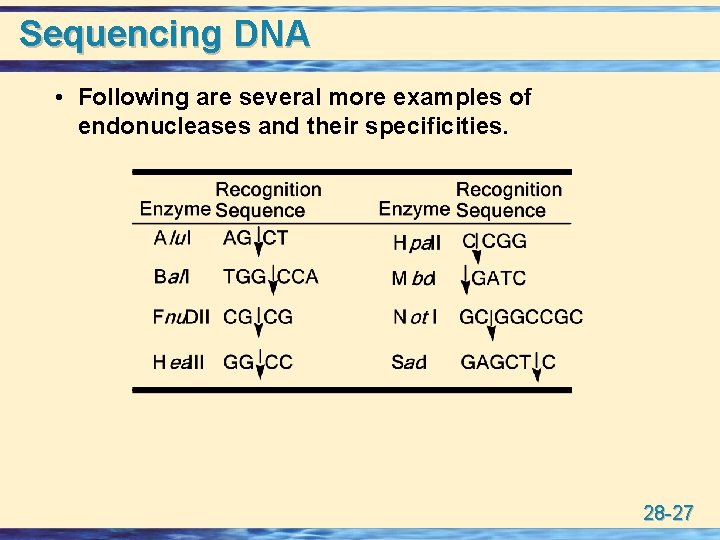
Sequencing DNA • Following are several more examples of endonucleases and their specificities. 28 -27

Sequencing DNA • Maxam-Gilbert method: A method developed by Allan Maxam and Walter Gilbert; depends on base-specific chemical cleavage. • Dideoxy chain termination method: Developed by Frederick Sanger. Gilbert and Sanger shared the 1980 Nobel Prize for biochemistry for their “development of chemical and biochemical analysis of DNA structure. ” 28 -28

Replication in Vitro • the sequence of nucleotides in one strand (ss. DNA) is copied as a complementary strand to form the second strand of a double-stranded DNA (ds. DNA). • Synthesis is catalyzed by DNA polymerase. • DNA polymerase requires • the four deoxynucleotide triphosphate (d. NTP) monomers • a primer is present to start the process. 28 -29

Dideoxy Chain Termination • Chain termination method is accomplished by the addition to the synthesizing medium of a 2’, 3’dideoxynucleotide triphosphate (dd. NTP). • Because a dd. NTP has no 3’-OH, chain synthesis is terminated when a dd. NTP becomes incorporated. Without a OH here chain cannot extend. 28 -30

Methodology of Dideoxy Chain Termination In this method, the following are mixed: • Single-stranded DNA of unknown sequence and primer; then divided into four reaction mixtures. u To each of the four reaction mixture is then added: • The four d. NTP, one of which is labeled in the 5’ end with phosphorus-32 which is radioactive. • DNA polymerase. • one of the four dd. NTPs. 28 -31

Dideoxy Chain Termination After gel electrophoresis of each reaction mixture • a piece of film is placed over the gel. • Gamma rays released by P-32 darken the film and create a pattern of the resolved oligonucleotide. • The base sequence of the complement to the original strand is read directly from bottom to top of the developed film. 28 -32

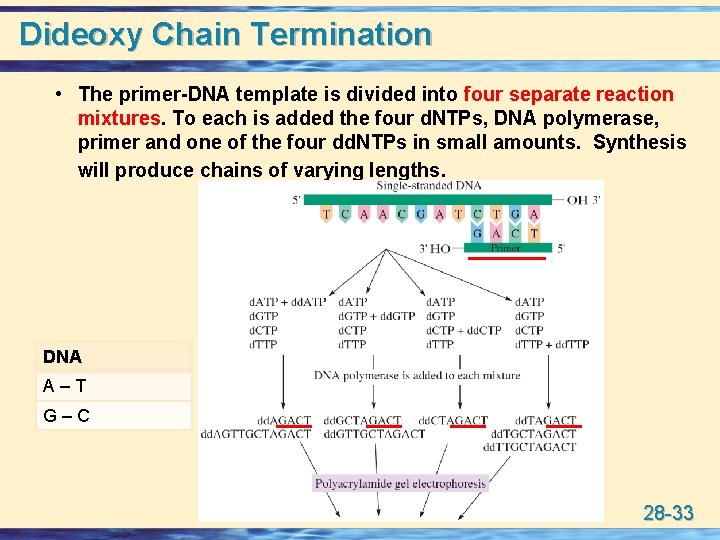
Dideoxy Chain Termination • The primer-DNA template is divided into four separate reaction mixtures. To each is added the four d. NTPs, DNA polymerase, primer and one of the four dd. NTPs in small amounts. Synthesis will produce chains of varying lengths. DNA A–T G–C 28 -33

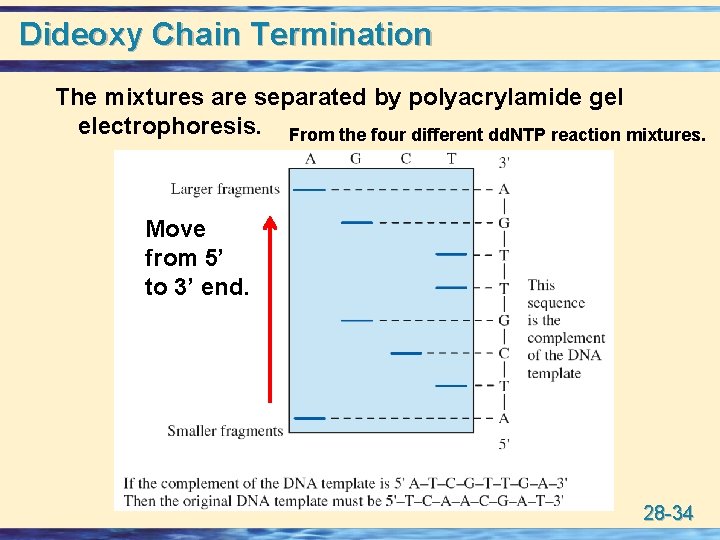
Dideoxy Chain Termination The mixtures are separated by polyacrylamide gel electrophoresis. From the four different dd. NTP reaction mixtures. Move from 5’ to 3’ end. 28 -34