Nucleic Acid Programmable Protein Array NAPPA as a

Nucleic Acid Programmable Protein Array (NAPPA) as a tool for diagnostics Samuel Kirkeng 7 December, 2017

What makes NAPPA special? Cell-free expression High-throughput Thousands of DNA sequences coding for proteins can be spotted simultaneously Less time consuming Lysate washed over slides provides material for coupled transcription/translation Purify only the DNA, instead of expressing then purifying the proteins themselves Printing DNA is more reliable Greater accuracy and

Basic Microarray Methodology Proteins marked with fluorescence Slide or chip is arrayed with different wells/washed with a coat that will immobilize, hold, protect, and orient proteins Epoxy or other material Expose binding sites, prevent the protein from denaturing Capture molecules antibodies Probe molecules Fluorescently labelled proteins Interactions between labeled proteins and immobilized protein results in fluorescence. Laser scanner analyzes the fluorescence
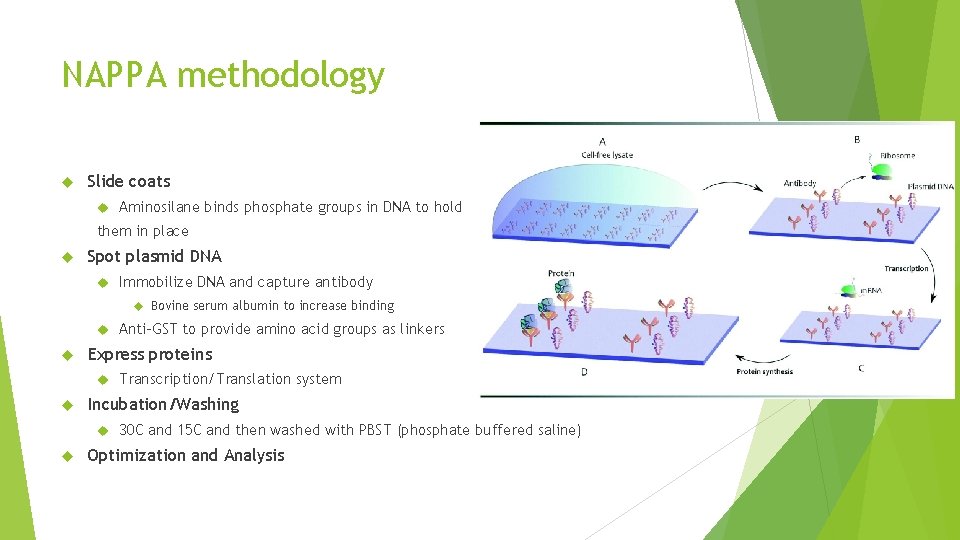
NAPPA methodology Slide coats Aminosilane binds phosphate groups in DNA to hold them in place Spot plasmid DNA Immobilize DNA and capture antibody Bovine serum albumin to increase binding Express proteins Transcription/Translation system Incubation/Washing Anti-GST to provide amino acid groups as linkers 30 C and 15 C and then washed with PBST (phosphate buffered saline) Optimization and Analysis
Case study: NAPPA in ovarian cancer research Identify the Tumor-associated autoantibodies (TAAb) biomarkers that could be used to tell cancerous patients from those that were healthy or had a benign form of the cancer TAAb= autoantibodies generated due to overexpression/mutation of proteins A few TAAbs and antigens against TAAbs have been identified in patients, but identifying more could lead to faster and more accurate diagnosis 735 antigen sites (to select against the TAAb markers), analysis of over 10, 000 human proteins between 30 healthy individuals, 30 individuals with benign ovarian cancer, and 30 with ovarian cancer
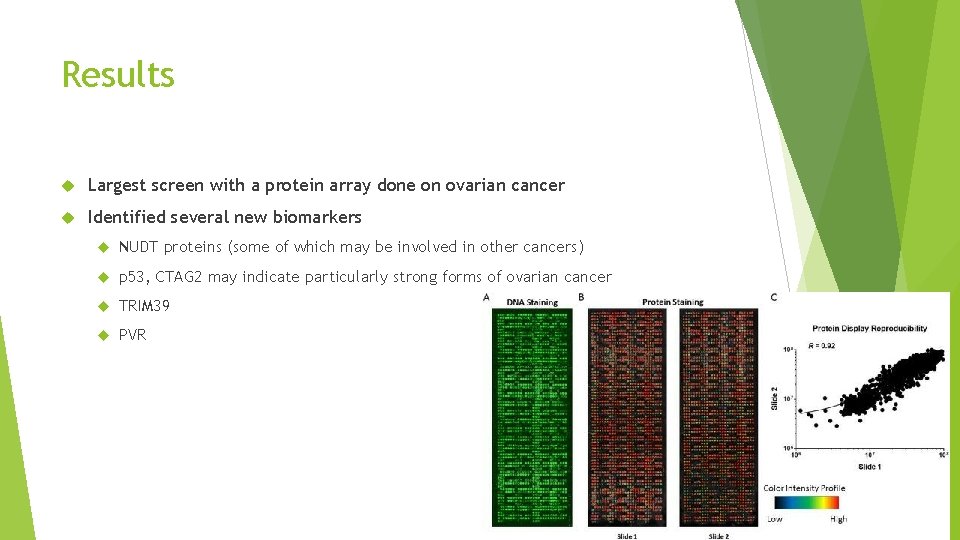
Results Largest screen with a protein array done on ovarian cancer Identified several new biomarkers NUDT proteins (some of which may be involved in other cancers) p 53, CTAG 2 may indicate particularly strong forms of ovarian cancer TRIM 39 PVR

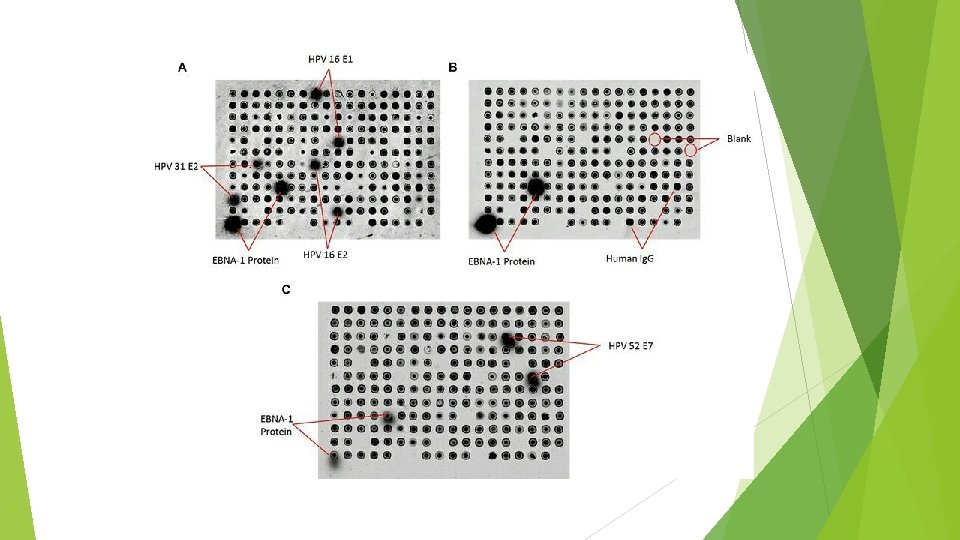
Case study: NAPPA in HPV-related cancer research (cervical cancer) 600, 000 cancers are related to Human Pampillomavirus each year The virus itself has a small double-stranded sequence, but there are over 100 forms of HPV In response to HPV cancers, an HPV-specific Ig. G antibody is produced
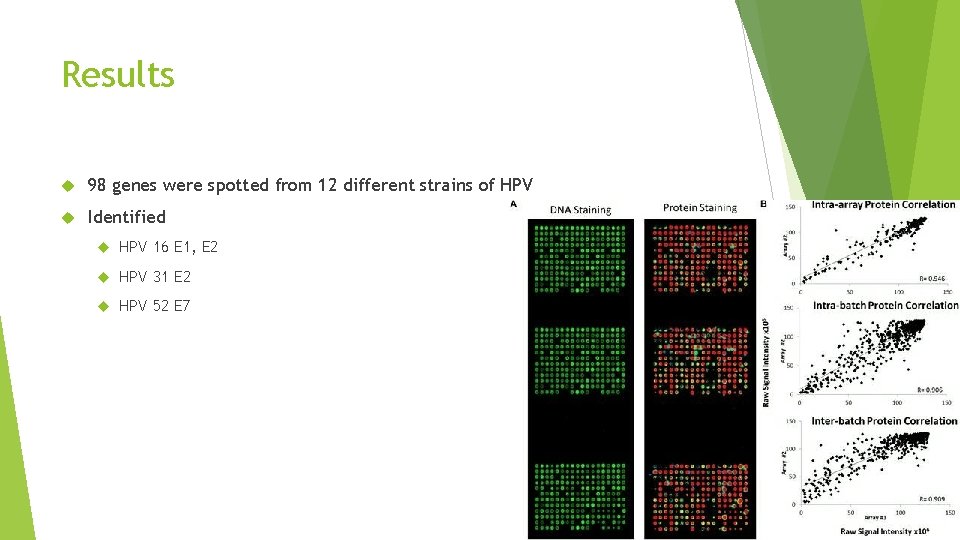
Results 98 genes were spotted from 12 different strains of HPV Identified HPV 16 E 1, E 2 HPV 31 E 2 HPV 52 E 7

Katchman, B. A. , Chomwell, D. , Wallstrom, G. , Vitonis, A. F. , La. Baer, J. , Cramer, D. W. , & Anderson, K. S. (July 2017). Autoantibody biomarkers for the detection of serous ovarian cancer. Gynecologic Oncology, 146(1), 129 -136. Retrieved November 28, 2017, from http: //www. sciencedirect. com. proxy. library. ohio. edu/science/article/pii/S 0090825817 307 758? via%3 Dihub Diez, P. , Gonzalez-Gonzalez, M. , Lourido, L. , Degano, R. M. , Ibarrola, N. , Casado. Vela, J. , Fuentes, M. (2015). NAPPA as a Real New Method for Protein Microarray Generation. Microarrays, 4(2), 214 -227. Retrieved December 1, 2017, from http: //www. mdpi. com/2076 -3905/4/2/214/htm Ewaisha, R. , Meshay, I. , Resnik, J. , Katchman, B. A. and Anderson, K. S. (2016), Programmable protein arrays for immunoprofiling HPV-associated cancers. Proteomics, 16: 1215– 1224. doi: 10. 1002/pmic. 201500376, from http: //onlinelibrary. wiley. com. proxy. library. ohio. edu/doi/10. 1002/pmic. 201500376/ful l
- Slides: 10