Nuclear receptor ligandbinding domains looked at from all

Nuclear receptor ligand-binding domains, looked at from all directions.
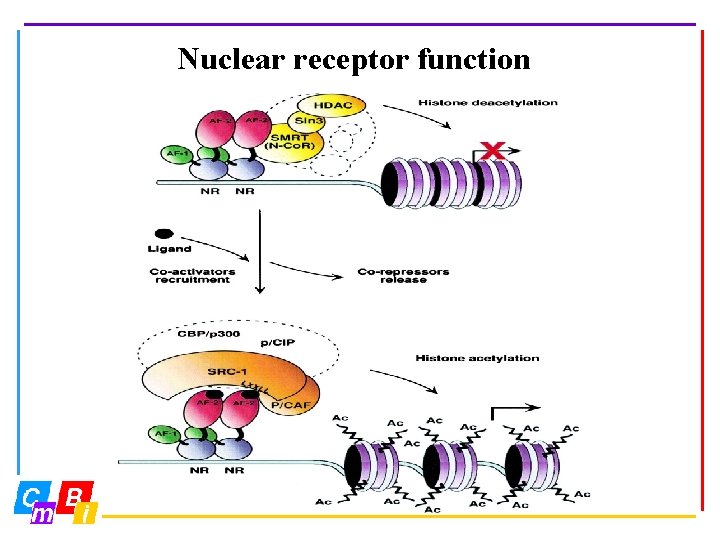
Nuclear receptor function
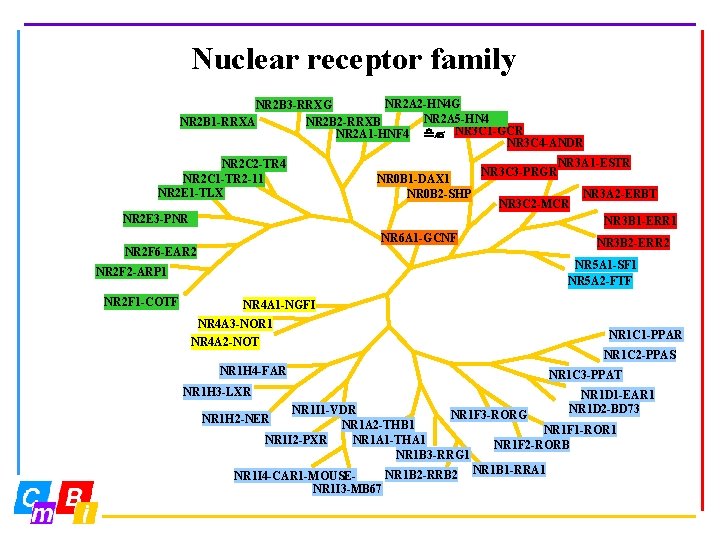
Nuclear receptor family NR 2 A 2 -HN 4 G NR 2 B 3 -RRXG NR 2 A 5 -HN 4 NR 2 B 1 -RRXA NR 2 B 2 -RRXB NR 2 A 1 -HNF 4 d? NR 3 C 1 -GCR NR 3 C 4 -ANDR NR 2 C 2 -TR 4 NR 2 C 1 -TR 2 -11 NR 2 E 1 -TLX NR 0 B 1 -DAX 1 NR 0 B 2 -SHP NR 2 E 3 -PNR NR 3 A 1 -ESTR NR 3 C 2 -MCR NR 3 A 2 -ERBT NR 3 B 1 -ERR 1 NR 6 A 1 -GCNF NR 2 F 6 -EAR 2 NR 3 B 2 -ERR 2 NR 5 A 1 -SF 1 NR 5 A 2 -FTF NR 2 F 2 -ARP 1 NR 2 F 1 -COTF NR 3 C 3 -PRGR NR 4 A 1 -NGFI NR 4 A 3 -NOR 1 NR 1 C 1 -PPAR NR 4 A 2 -NOT NR 1 C 2 -PPAS NR 1 H 4 -FAR NR 1 C 3 -PPAT NR 1 H 3 -LXR NR 1 D 1 -EAR 1 NR 1 D 2 -BD 73 NR 1 I 1 -VDR NR 1 F 3 -RORG NR 1 A 2 -THB 1 NR 1 F 1 -ROR 1 NR 1 I 2 -PXR NR 1 A 1 -THA 1 NR 1 F 2 -RORB NR 1 B 3 -RRG 1 NR 1 B 2 -RRB 2 NR 1 B 1 -RRA 1 NR 1 I 4 -CAR 1 -MOUSE- NR 1 H 2 -NER NR 1 I 3 -MB 67
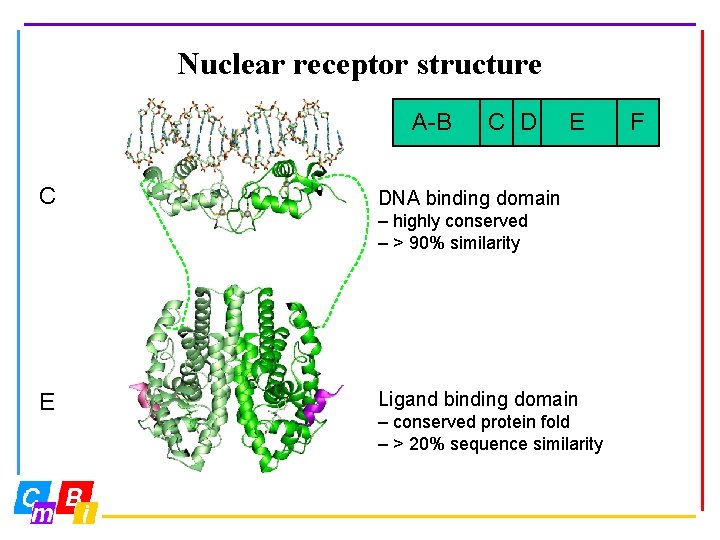
Nuclear receptor structure A-B AF-1 C C D DNA E LBD DNA binding domain – highly conserved – > 90% similarity E Ligand binding domain – conserved protein fold – > 20% sequence similarity F

The questions As Organon is paying the bills, question one is, of course☺, how do ligands relate to activity? NRs can bind co-activators and co-repressors, with or without ligand being present, so what are agonists, antagonists, and inverse agonists? What is the role of each amino acid in the NR LBD? Which data handling is needed to answer these questions?

3 D structure LBD (h. ER )
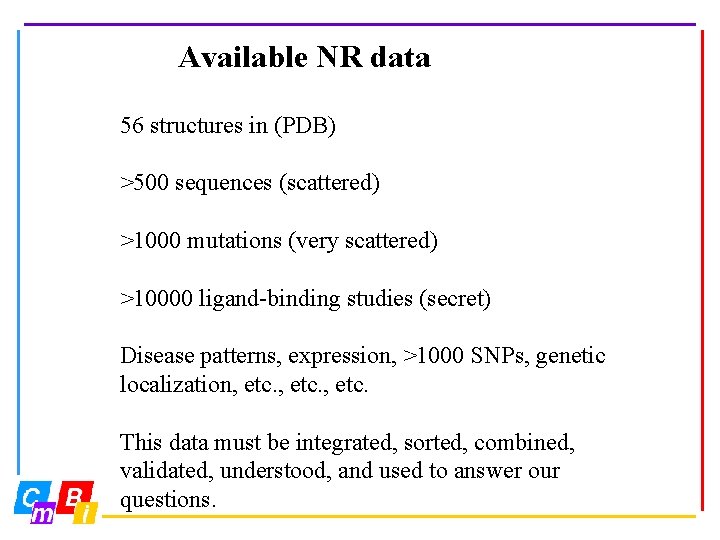
Available NR data 56 structures in (PDB) >500 sequences (scattered) >1000 mutations (very scattered) >10000 ligand-binding studies (secret) Disease patterns, expression, >1000 SNPs, genetic localization, etc. This data must be integrated, sorted, combined, validated, understood, and used to answer our questions.

Step 1 The first important step is a common numbering scheme. Whoever solves that problem once and for all should get three Nobel prices.
Large data volumes allow us to develop new data analysis techniques. Entropy-variability analysis is a novel technique to look at very large multiple sequence alignments. Entropy-variability analysis requires ‘better’ alignments than routinely are obtained with ‘standard’ multiple sequence alignment programs.
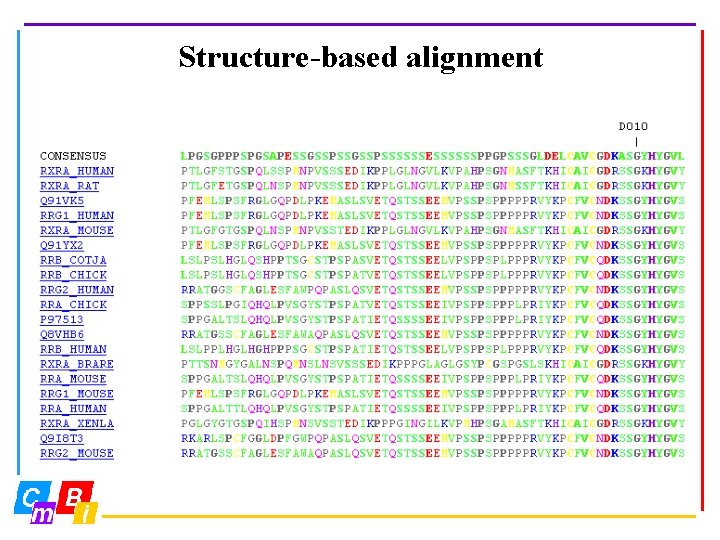
Structure-based alignment
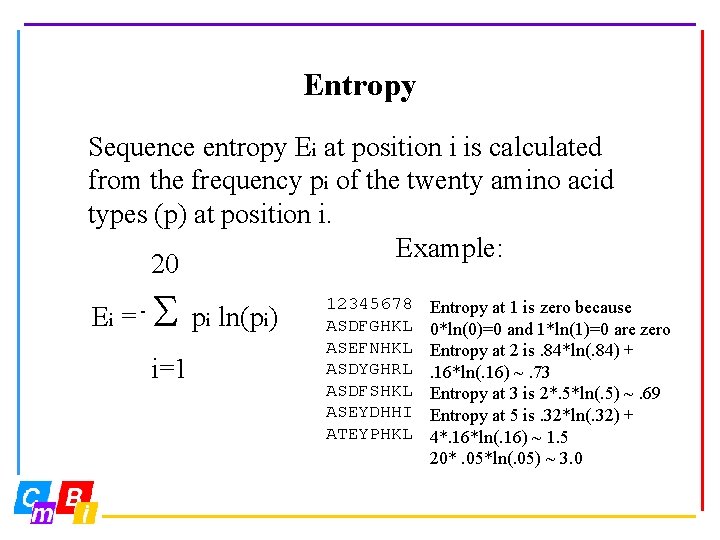
Entropy Sequence entropy Ei at position i is calculated from the frequency pi of the twenty amino acid types (p) at position i. Example: 20 Ei = - S i=1 pi ln(pi) 12345678 ASDFGHKL ASEFNHKL ASDYGHRL ASDFSHKL ASEYDHHI ATEYPHKL Entropy at 1 is zero because 0*ln(0)=0 and 1*ln(1)=0 are zero Entropy at 2 is. 84*ln(. 84) +. 16*ln(. 16) ~. 73 Entropy at 3 is 2*. 5*ln(. 5) ~. 69 Entropy at 5 is. 32*ln(. 32) + 4*. 16*ln(. 16) ~ 1. 5 20*. 05*ln(. 05) ~ 3. 0
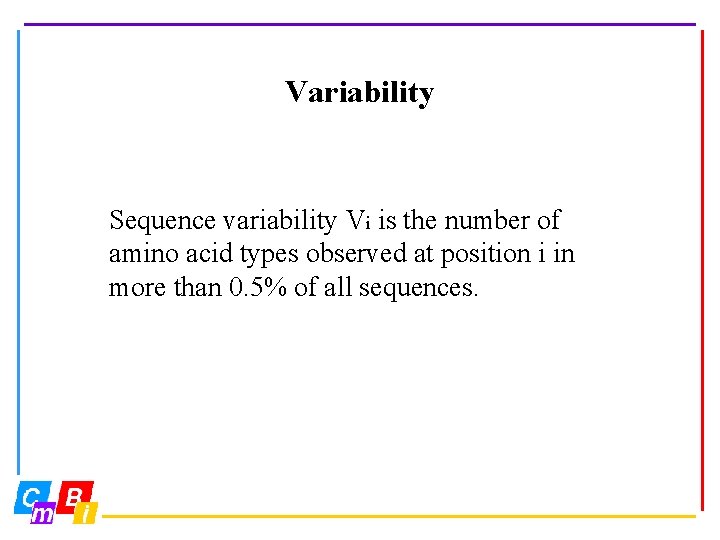
Variability Sequence variability Vi is the number of amino acid types observed at position i in more than 0. 5% of all sequences.
Rules 1) If a residue is conserved, 2) it is important 2) If a residue is very conserved, it is very important
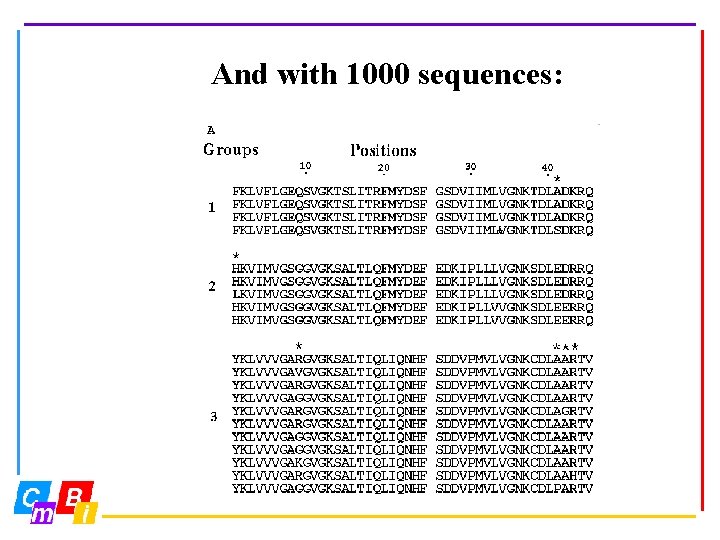
And with 1000 sequences:
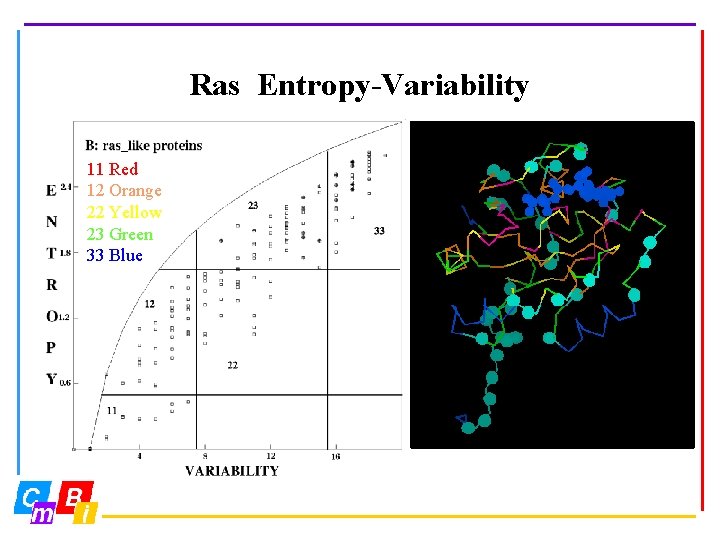
Ras Entropy-Variability 11 Red 12 Orange 22 Yellow 23 Green 33 Blue
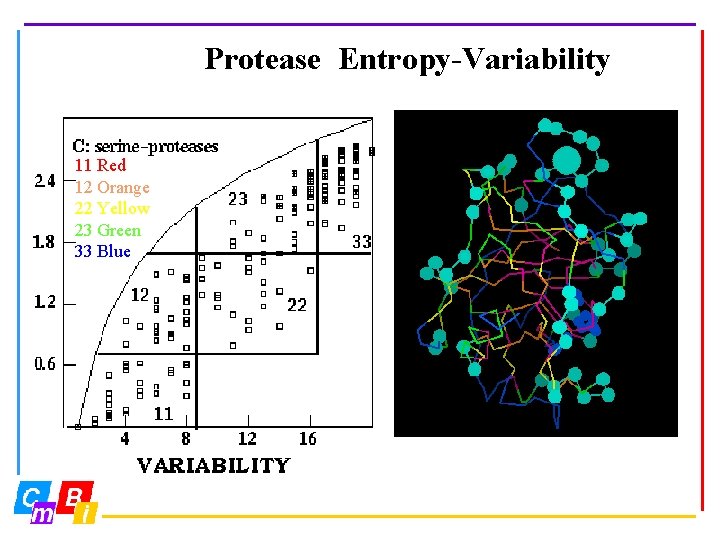
Protease Entropy-Variability 11 Red 12 Orange 22 Yellow 23 Green 33 Blue
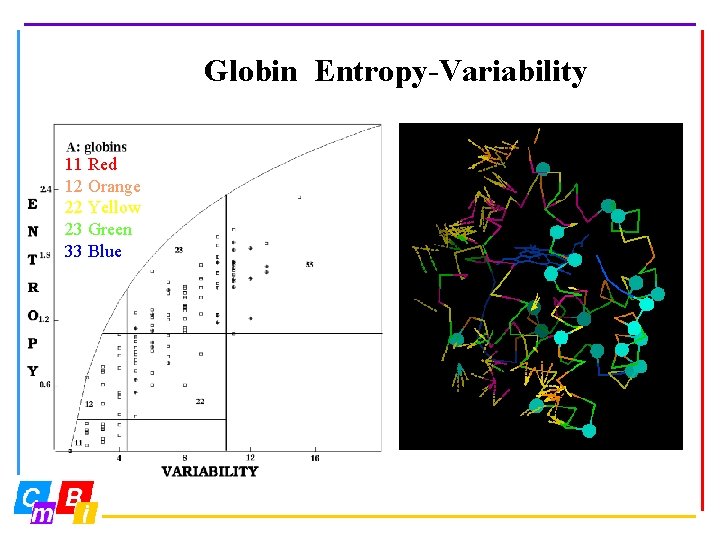
Globin Entropy-Variability 11 Red 12 Orange 22 Yellow 23 Green 33 Blue
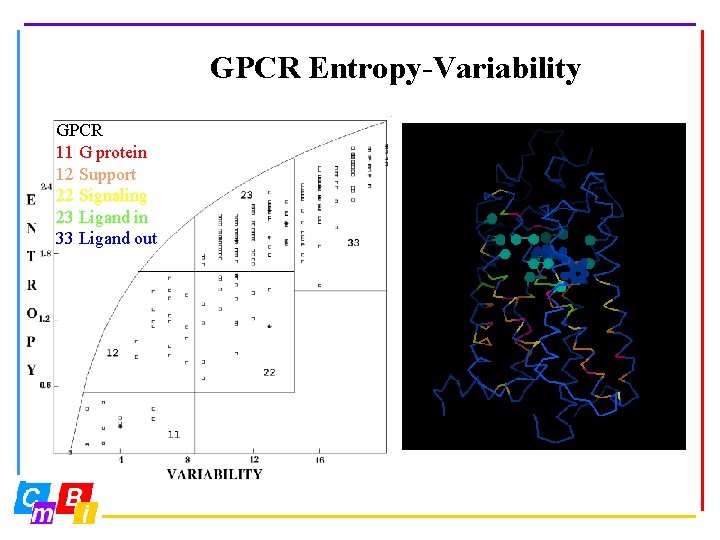
GPCR Entropy-Variability GPCR 11 G protein 12 Support 22 Signaling 23 Ligand in 33 Ligand out
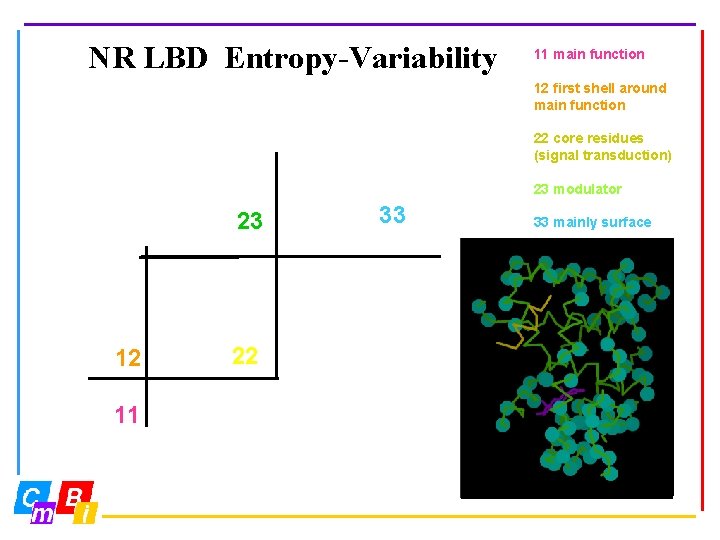
NR LBD Entropy-Variability 11 main function 12 first shell around main function 22 core residues (signal transduction) 23 modulator 23 12 11 22 33 33 mainly surface
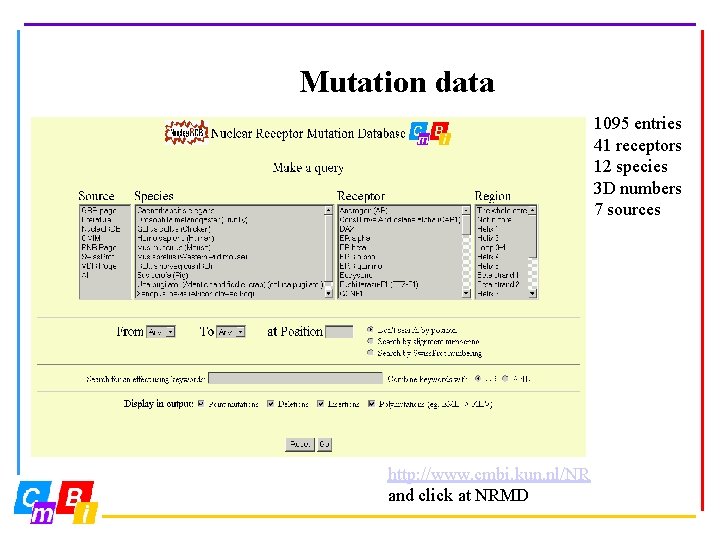
Mutation data 1095 entries 41 receptors 12 species 3 D numbers 7 sources http: //www. cmbi. kun. nl/NR and click at NRMD
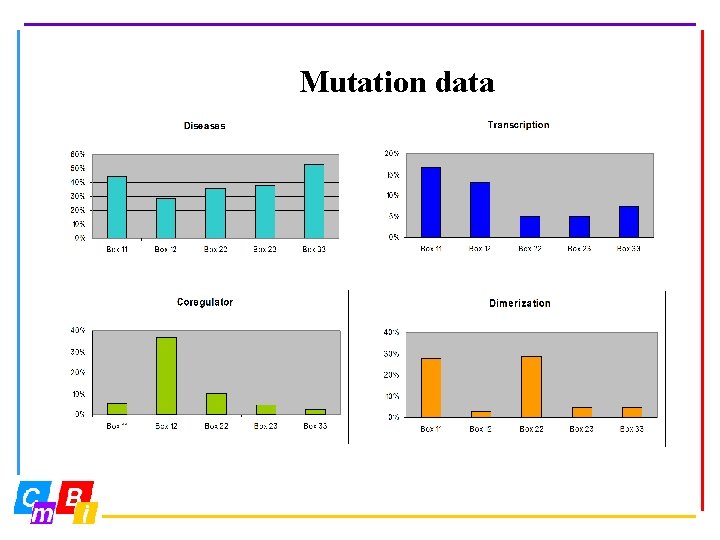
Mutation data
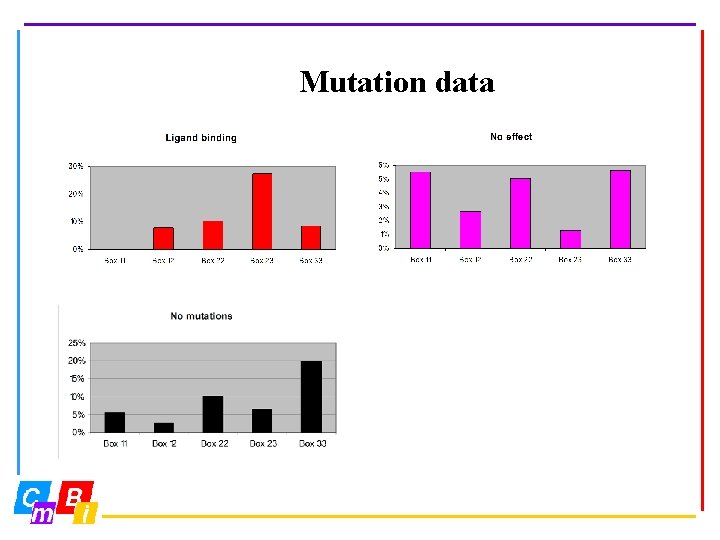
Mutation data
Ligand binding data Ligand-binding positions extracted from PDB files (nomenclature) Categorized in very frequent to not so frequent binder Which type of ligand it binds (agonist/antagonist=inverse agonist…)
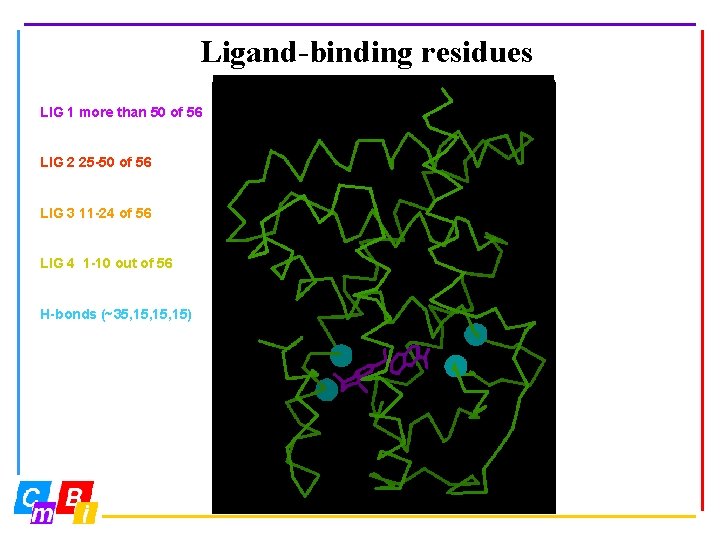
Ligand-binding residues LIG 1 more than 50 of 56 LIG 2 25 -50 of 56 LIG 3 11 -24 of 56 LIG 4 1 -10 out of 56 H-bonds (~35, 15, 15)
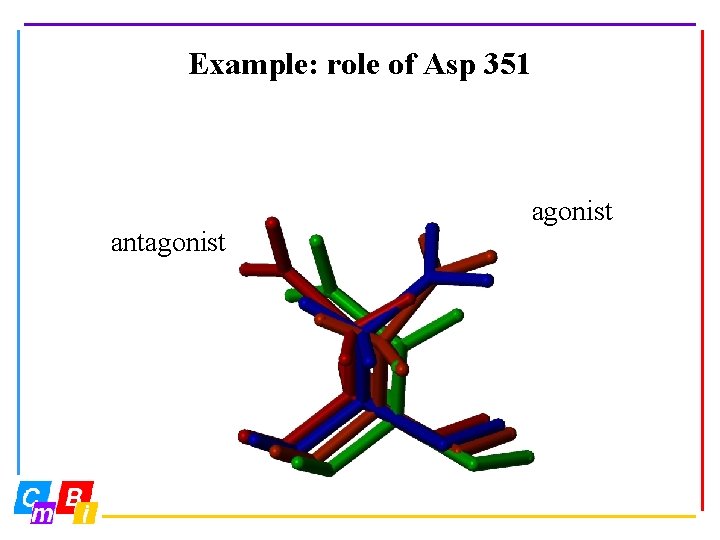
Example: role of Asp 351 agonist antagonist
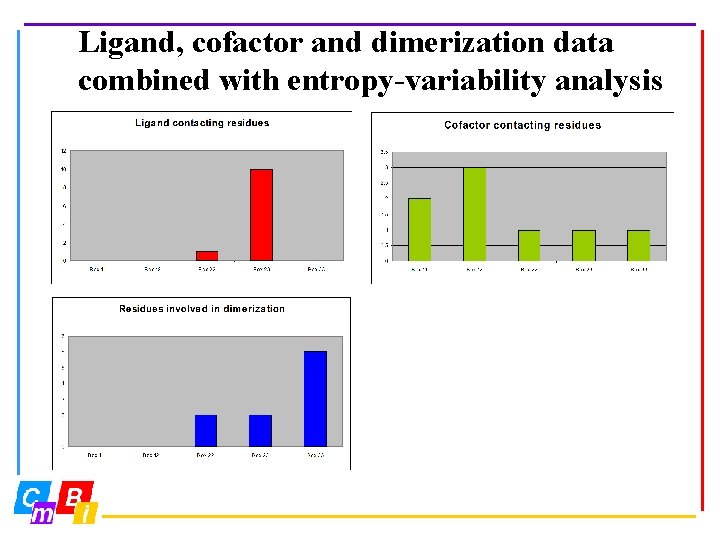
Ligand, cofactor and dimerization data combined with entropy-variability analysis
Conclusions: Data is difficult, but we need it (sic); life would be so nice if we could do without. PDB files are the worst. Nomenclature is not homogeneous. Much data has been carefully hidden in the literature where it can only be found back with great difficulty. Residue numbering is difficult but very necessary. Variability-entropy analysis is powerful, but requires very 'good' alignments.

Acknowledgements: Organon Jacob de Vlieg Jan Klomp Paula van Noort Scott Lusher UCSF Florence Horn CMBI Emmanuel Bettler Simon Folkertsma Henk-Jan Joosten Joost van Durme Wilco Fleuren Jeroen Eitjes Jeroen van Broekhuizen Richard Notebaart Richard van Hameren Ralph Brandt
- Slides: 28