Novel Prochlorococcus ecotypes from the Anoxic Minimum Zone





- Slides: 23

Novel Prochlorococcus ecotypes from the Anoxic Minimum Zone of the Eastern Tropical South Pacific Ocean. Alvaro Munoz Plominsky

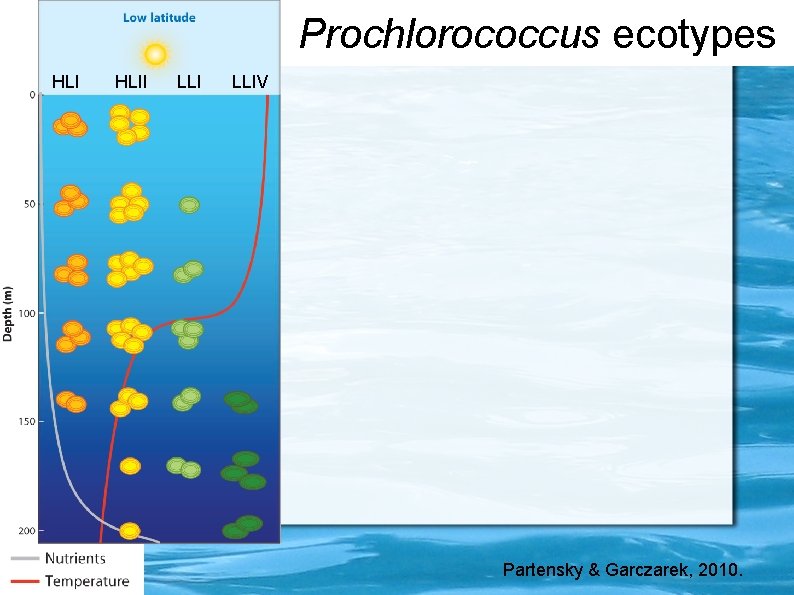
Prochlorococcus ecotypes HLII LLIV Partensky & Garczarek, 2010.

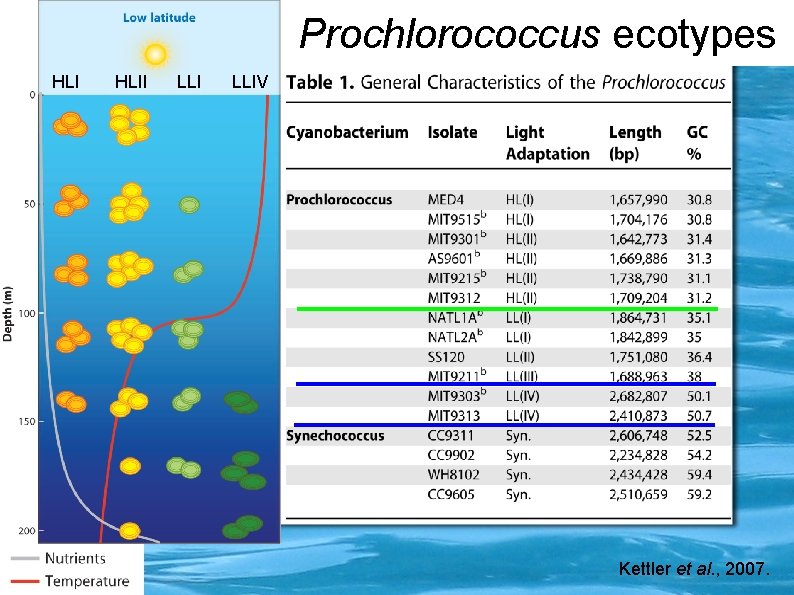
Prochlorococcus ecotypes HLII LLIV Kettler et al. , 2007.

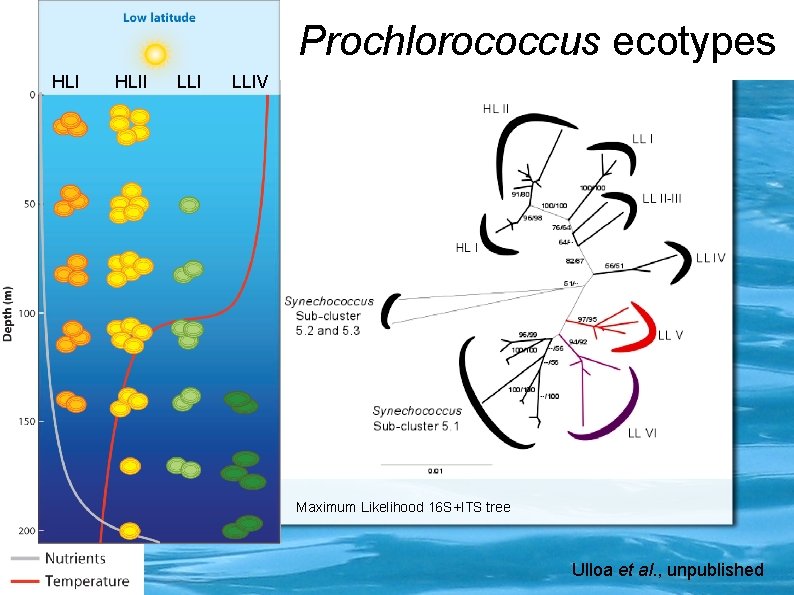
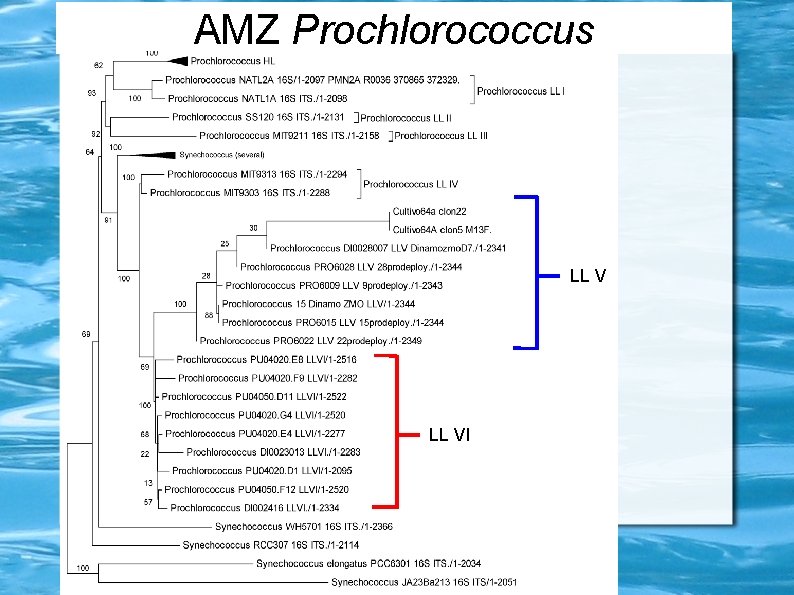
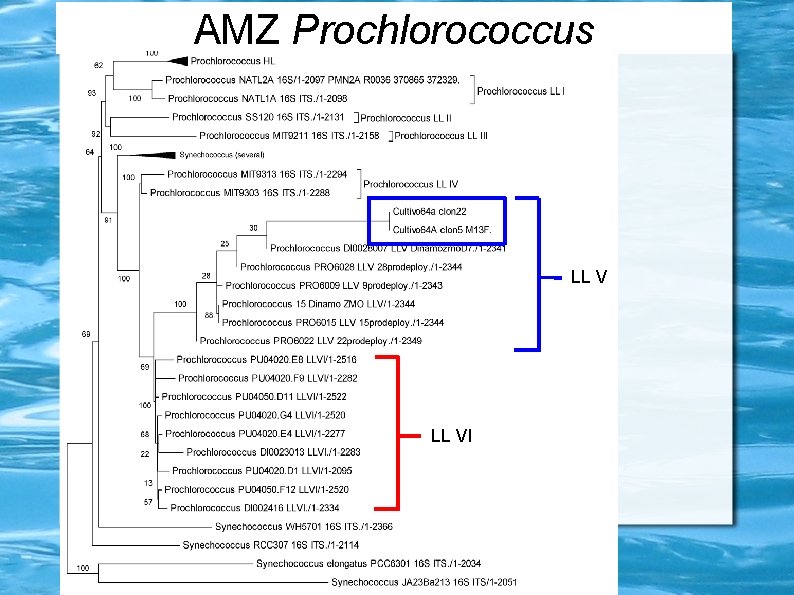
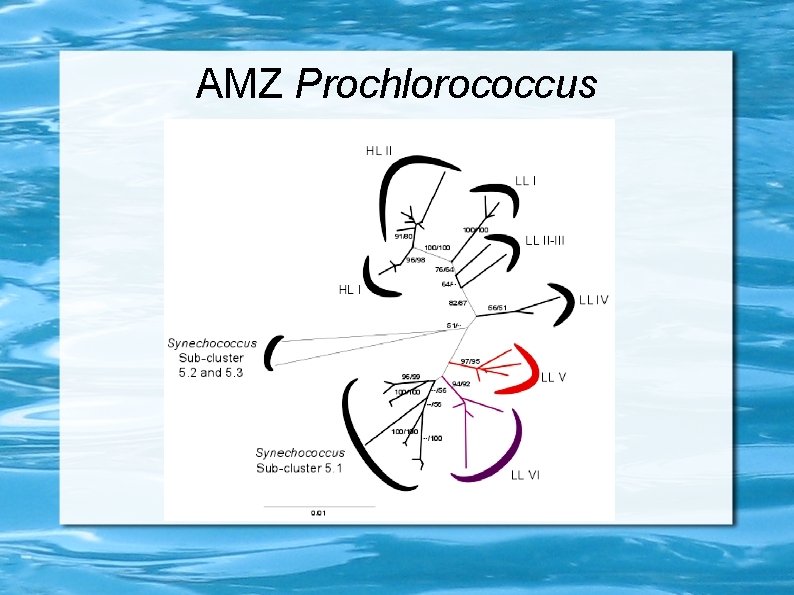
Prochlorococcus ecotypes HLII LLIV Maximum Likelihood 16 S+ITS tree Ulloa et al. , unpublished

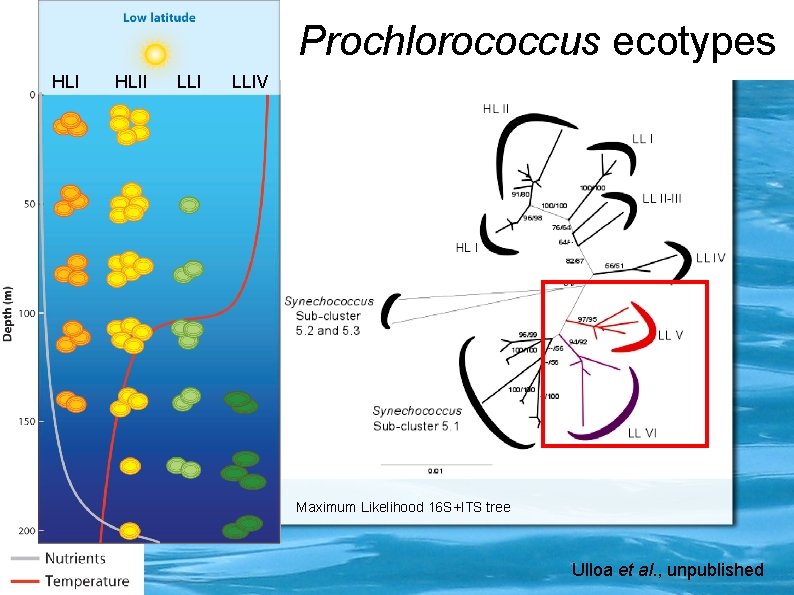
Prochlorococcus ecotypes HLII LLIV Maximum Likelihood 16 S+ITS tree Ulloa et al. , unpublished

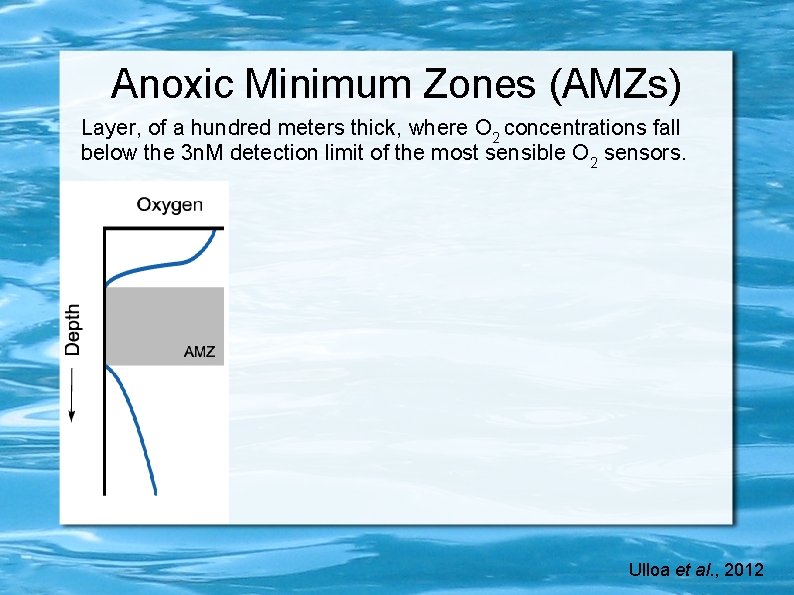
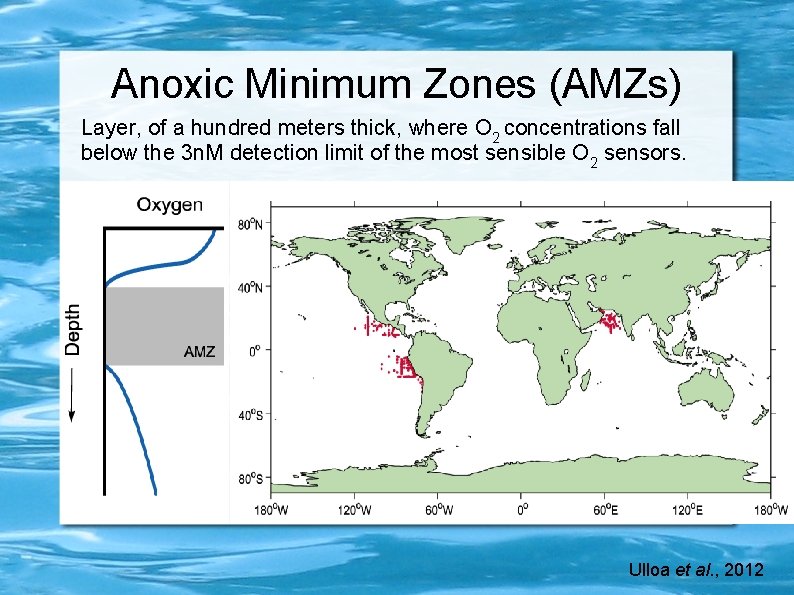
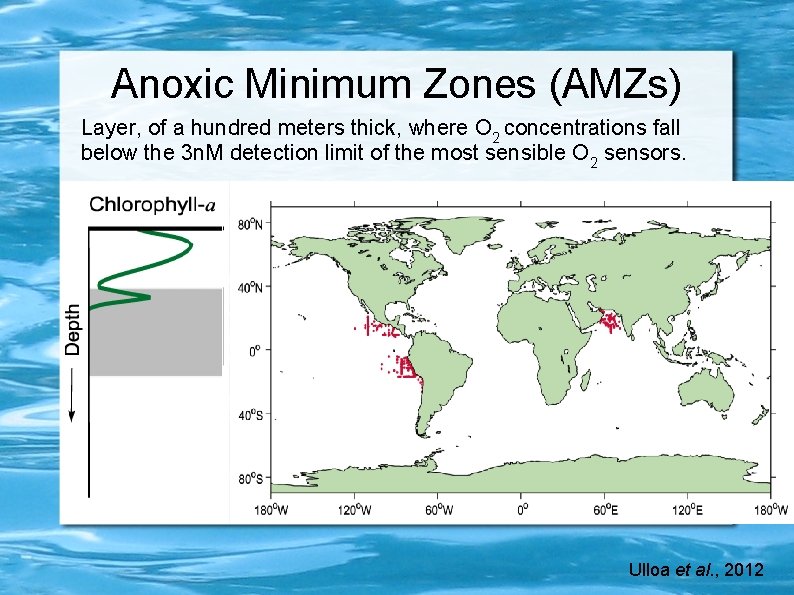
Anoxic Minimum Zones (AMZs) Layer, of a hundred meters thick, where O 2 concentrations fall below the 3 n. M detection limit of the most sensible O 2 sensors. Ulloa et al. , 2012

Anoxic Minimum Zones (AMZs) Layer, of a hundred meters thick, where O 2 concentrations fall below the 3 n. M detection limit of the most sensible O 2 sensors. Ulloa et al. , 2012

Anoxic Minimum Zones (AMZs) Layer, of a hundred meters thick, where O 2 concentrations fall below the 3 n. M detection limit of the most sensible O 2 sensors. Ulloa et al. , 2012

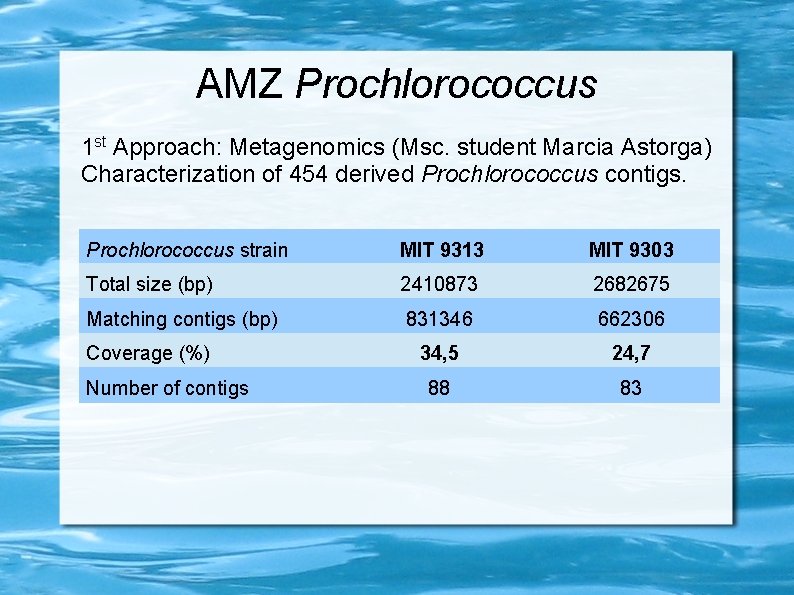
AMZ Prochlorococcus 1 st Approach: Metagenomics (Msc. student Marcia Astorga) Characterization of 454 derived Prochlorococcus contigs. Prochlorococcus strain MIT 9313 MIT 9303 Total size (bp) 2410873 2682675 Matching contigs (bp) 831346 662306 34, 5 24, 7 88 83 Coverage (%) Number of contigs

AMZ Prochlorococcus 1 st Approach: Metagenomics (Msc. student Marcia Astorga) Characterization of 454 derived Prochlorococcus contigs. 2 nd Approach: Single-Cell Genome Sequencing

AMZ Prochlorococcus 2 nd Approach: Single-Cell Genome Sequencing

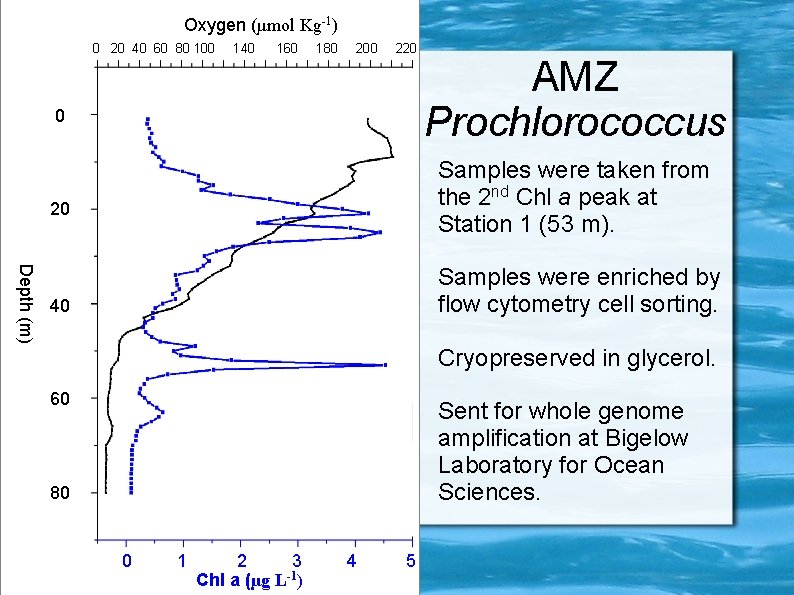
Oxygen (µmol Kg-1) 0 20 40 60 80 100 140 160 180 200 220 0 AMZ Prochlorococcus Samples were taken from the 2 nd Chl a peak at Station 1 (53 m). 20 Depth (m) Samples were enriched by flow cytometry cell sorting. 40 Cryopreserved in glycerol. 60 Sent for whole genome amplification at Bigelow Laboratory for Ocean Sciences. 80 0 1 2 3 Chl a (µg L-1) 4 5

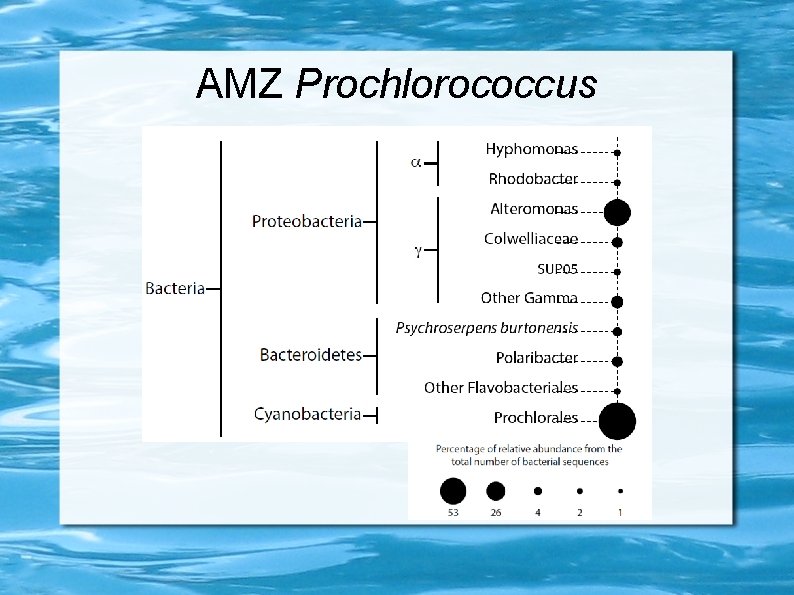
AMZ Prochlorococcus

AMZ Prochlorococcus 1 st Approach: Metagenomics (Msc. student Marcia Astorga) Characterization of 454 derived Prochlorococcus contigs. 2 nd Approach: Single-Cell Genome Sequencing 3 rd Approach: Culture Screening by q. PCR Approx. 350 cultures (of 15 -20 m. L), from Big-RAPA Station 1, were maintained in our laboratory!!

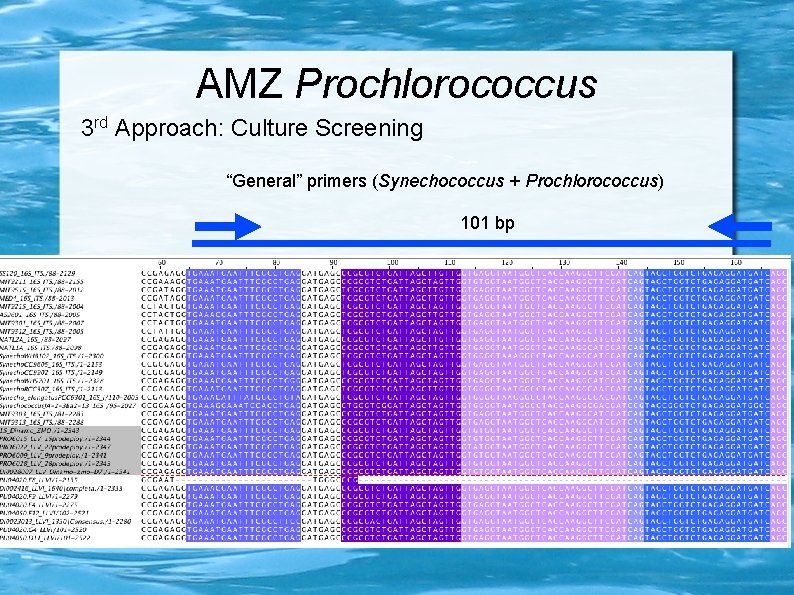
AMZ Prochlorococcus 3 rd Approach: Culture Screening “General” primers (Synechococcus + Prochlorococcus) 101 bp

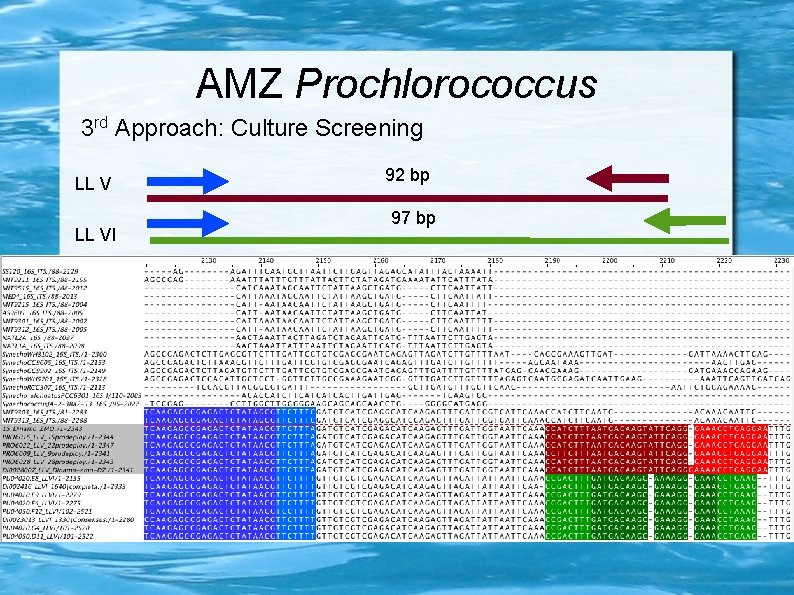
AMZ Prochlorococcus 3 rd Approach: Culture Screening LL VI 92 bp 97 bp

AMZ Prochlorococcus 3 rd Approach: Culture Screening LL VI

AMZ Prochlorococcus 3 rd Approach: Culture Screening LL VI

AMZ Prochlorococcus Single-cell genomes will be available. . . eventually. Genomic data of AMZ Prochlorococcus is also available among metagenomic datasets. Cultures are currently being scaled for future isolation of pure LL V Procholococcus. q. PCR methodology will allow screening of samples from the 2013 cruises to the ETNP and ETSP OMZs.

AMZ Prochlorococcus

Lavin et al. , 2010.

