NOVEL PARADIGMS FOR DRUG DISCOVERY SHOTGUN COMPUTATIONAL MULTITARGET







- Slides: 18

NOVEL PARADIGMS FOR DRUG DISCOVERY SHOTGUN COMPUTATIONAL MULTITARGET SCREENING RAM SAMUDRALA ASSOCIATE PROFESSOR UNIVERSITY OF WASHINGTON

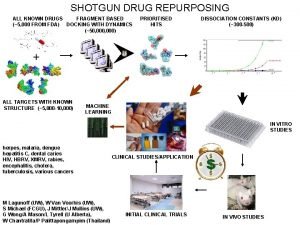
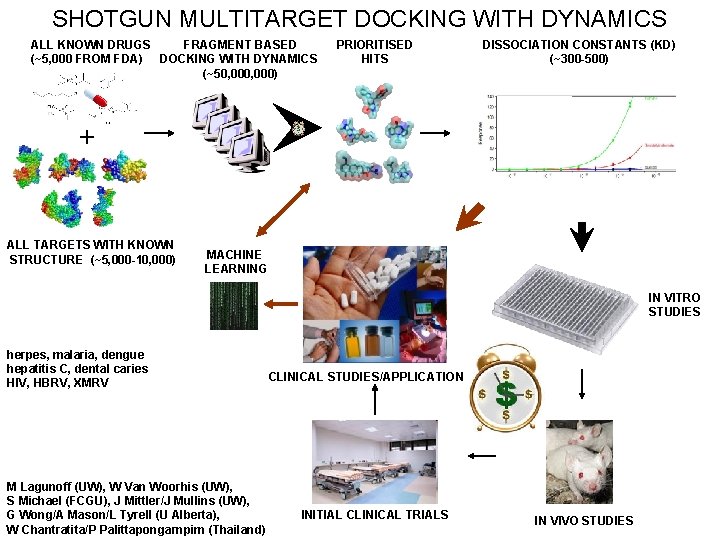
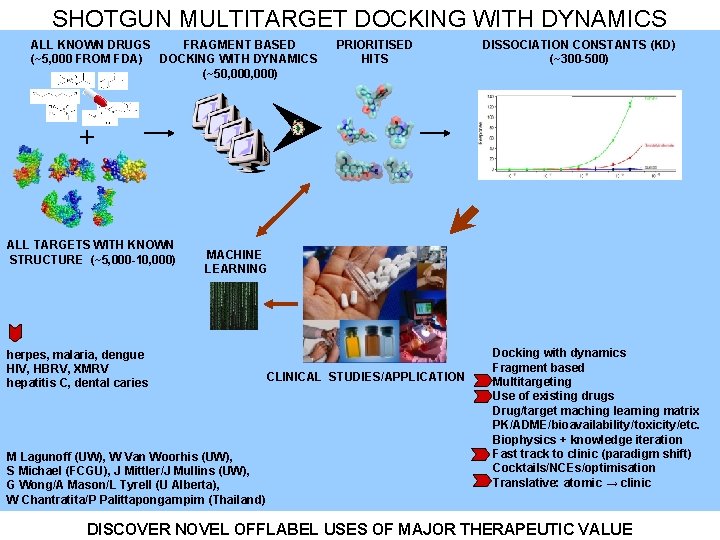
SHOTGUN MULTITARGET DOCKING WITH DYNAMICS ALL KNOWN DRUGS FRAGMENT BASED (~5, 000 FROM FDA) DOCKING WITH DYNAMICS (~50, 000) PRIORITISED HITS DISSOCIATION CONSTANTS (KD) (~300 -500) + ALL TARGETS WITH KNOWN STRUCTURE (~5, 000 -10, 000) MACHINE LEARNING IN VITRO STUDIES herpes, malaria, dengue hepatitis C, dental caries HIV, HBRV, XMRV M Lagunoff (UW), W Van Woorhis (UW), S Michael (FCGU), J Mittler/J Mullins (UW), G Wong/A Mason/L Tyrell (U Alberta), W Chantratita/P Palittapongarnpim (Thailand) CLINICAL STUDIES/APPLICATION INITIAL CLINICAL TRIALS IN VIVO STUDIES

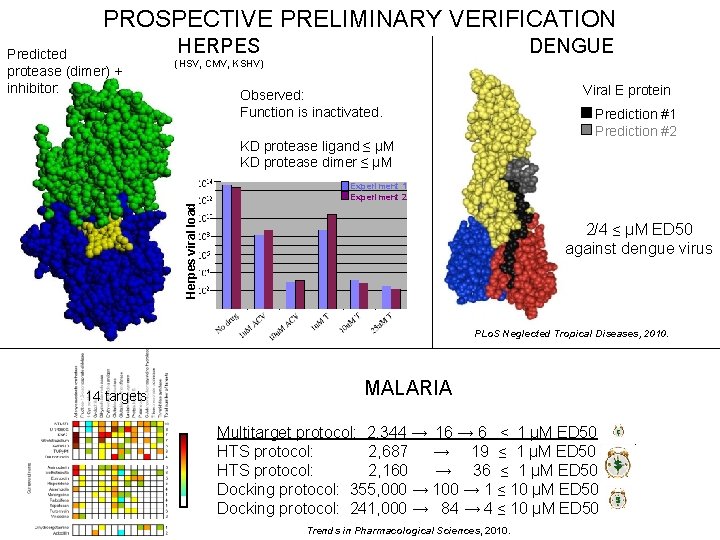
PROSPECTIVE PRELIMINARY VERIFICATION Predicted protease (dimer) + inhibitor: HERPES DENGUE (HSV, CMV, KSHV) Viral E protein Observed: Function is inactivated. Prediction #1 Prediction #2 KD protease ligand ≤ μM KD protease dimer ≤ μM Herpes viral load Experiment 1 Experiment 2 2/4 ≤ µM ED 50 against dengue virus PLo. S Neglected Tropical Diseases, 2010. 14 targets MALARIA Multitarget protocol: 2, 344 → 16 → 6 ≤ 1 µM ED 50 HTS protocol: 2, 687 → 19 ≤ 1 µM ED 50 HTS protocol: 2, 160 → 36 ≤ 1 µM ED 50 Docking protocol: 355, 000 → 1 ≤ 10 µM ED 50 Docking protocol: 241, 000 → 84 → 4 ≤ 10 µM ED 50 Trends in Pharmacological Sciences, 2010.

SHOTGUN MULTITARGET DOCKING WITH DYNAMICS ALL KNOWN DRUGS FRAGMENT BASED (~5, 000 FROM FDA) DOCKING WITH DYNAMICS (~50, 000) PRIORITISED HITS DISSOCIATION CONSTANTS (KD) (~300 -500) + ALL TARGETS WITH KNOWN STRUCTURE (~5, 000 -10, 000) MACHINE LEARNING herpes, malaria, dengue HIV, HBRV, XMRV hepatitis C, dental caries M Lagunoff (UW), W Van Woorhis (UW), S Michael (FCGU), J Mittler/J Mullins (UW), G Wong/A Mason/L Tyrell (U Alberta), W Chantratita/P Palittapongarnpim (Thailand) CLINICAL STUDIES/APPLICATION Docking with dynamics Fragment based Multitargeting Use of existing drugs Drug/target maching learning matrix PK/ADME/bioavailability/toxicity/etc. Biophysics + knowledge iteration Fast track to clinic (paradigm shift) Cocktails/NCEs/optimisation Translative: atomic → clinic DISCOVER NOVEL OFFLABEL USES OF MAJOR THERAPEUTIC VALUE


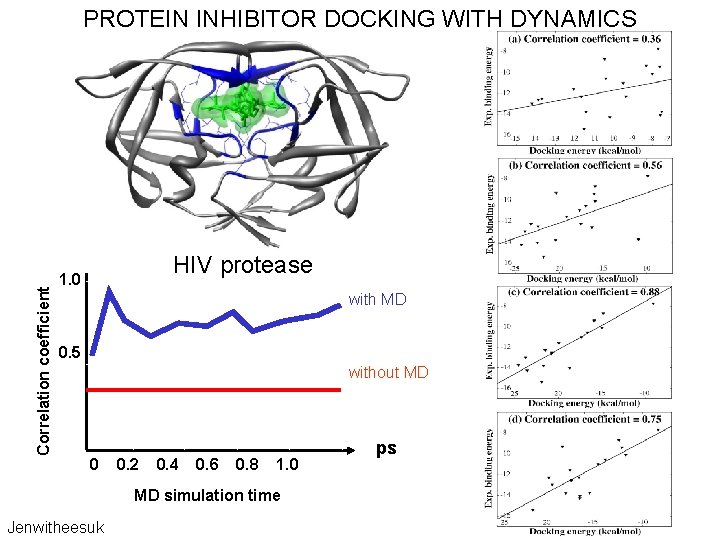
Correlation coefficient PROTEIN INHIBITOR DOCKING WITH DYNAMICS HIV protease 1. 0 with MD 0. 5 without MD 0 0. 2 0. 4 0. 6 0. 8 1. 0 MD simulation time Jenwitheesuk ps

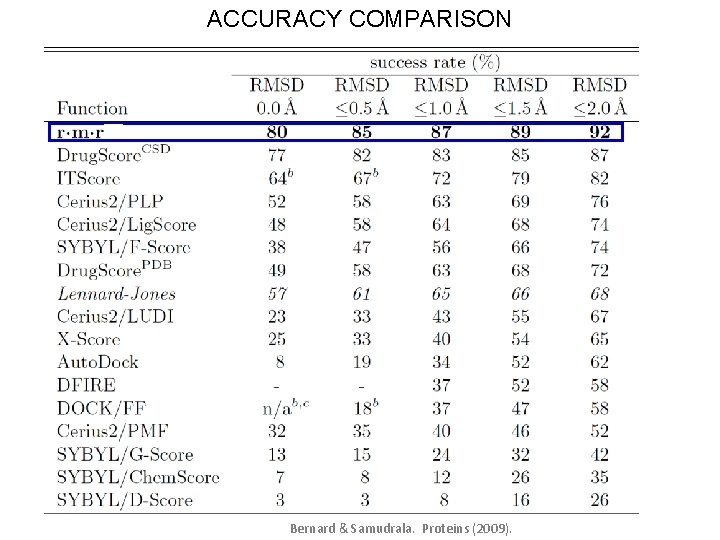
ACCURACY COMPARISON Bernard & Samudrala. Proteins (2009).

BACKGROUND AND MOTIVATION My research on protein and proteome structure, function, and interaction is directed to understanding how genomes specify phenotype and behaviour; my goal is to use this information to improve human health and quality of life. Protein functions and interactions are mediated by atomic three dimensional structure. We are applying all our structure prediction technologies to the area of small molecule therapeutic discovery. The goal is to create a comprehensive in silico drug discovery pipeline to increase the odds of initial preclinical hits and leads leading to significantly better outcomes downstream in the clinic. The knowledge-based drug discovery pipeline will adopt a shotgun approach that screens all known FDA approved drug and drug-like compounds against all known target proteins of known structure, simultaneously examining how a small molecule therapeutic interacts with targets, antitargets, metabolic pathways, to obtain a holistic picture of drug efficacy and side effects. Find new uses for existing drugs that can be used in the clinic, with a focus on third world and neglected diseases with poor or nonexisting treatments.

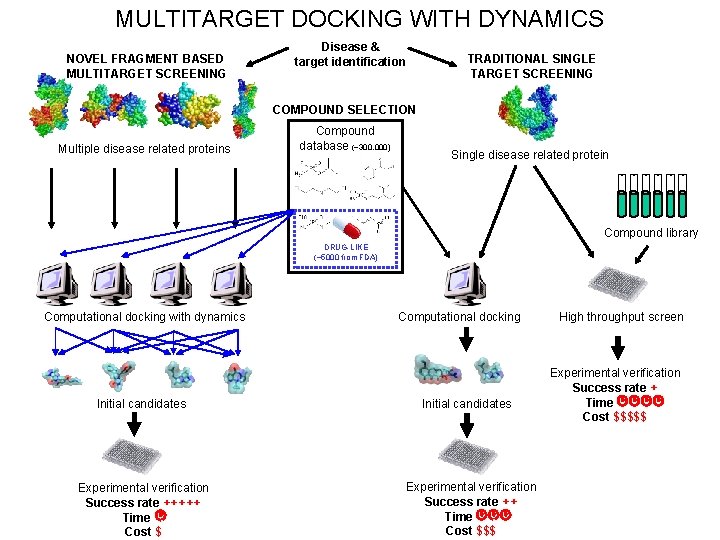
MULTITARGET DOCKING WITH DYNAMICS NOVEL FRAGMENT BASED MULTITARGET SCREENING Disease & target identification TRADITIONAL SINGLE TARGET SCREENING COMPOUND SELECTION Multiple disease related proteins Compound database (~300, 000) Single disease related protein Compound library DRUG-LIKE (~5000 from FDA) Computational docking with dynamics Initial candidates Experimental verification Success rate +++++ Time. Cost $ Computational docking Initial candidates Experimental verification Success rate ++ Time. Cost $$$ High throughput screen Experimental verification Success rate + Time. Cost $$$$$

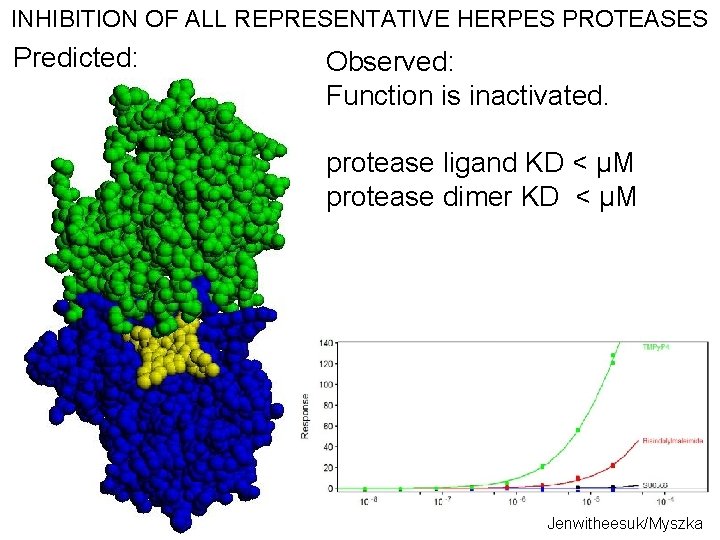
INHIBITION OF ALL REPRESENTATIVE HERPES PROTEASES Predicted: Observed: Function is inactivated. protease ligand KD < μM protease dimer KD < μM Jenwitheesuk/Myszka

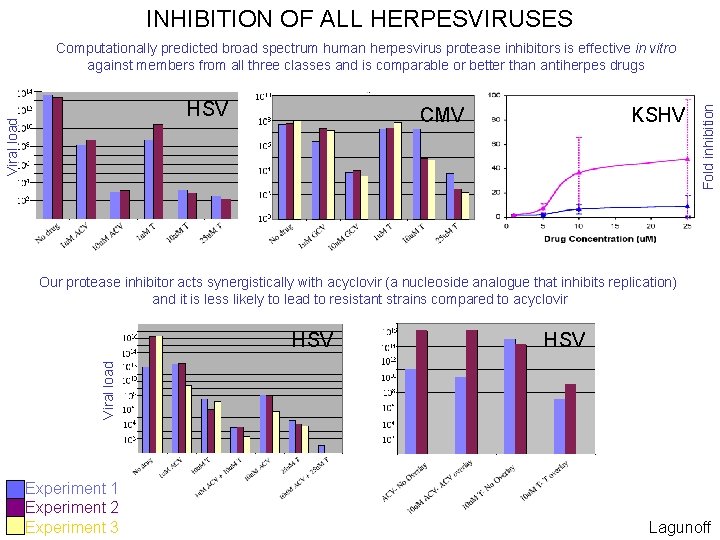
INHIBITION OF ALL HERPESVIRUSES Viral load HSV KSHV CMV Fold inhibition Computationally predicted broad spectrum human herpesvirus protease inhibitors is effective in vitro against members from all three classes and is comparable or better than antiherpes drugs Our protease inhibitor acts synergistically with acyclovir (a nucleoside analogue that inhibits replication) and it is less likely to lead to resistant strains compared to acyclovir HSV Viral load HSV Experiment 1 Experiment 2 Experiment 3 Lagunoff

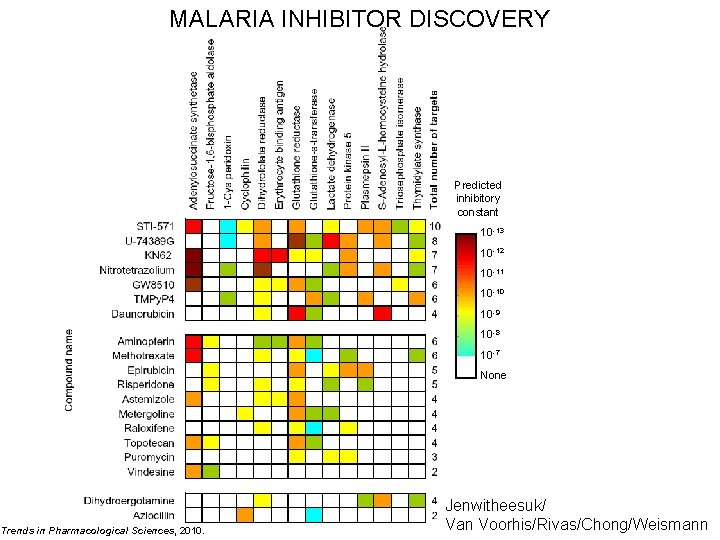
MALARIA INHIBITOR DISCOVERY Predicted inhibitory constant 10 -13 10 -12 10 -11 10 -10 10 -9 10 -8 10 -7 None Trends in Pharmacological Sciences, 2010. Jenwitheesuk/ Van Voorhis/Rivas/Chong/Weismann

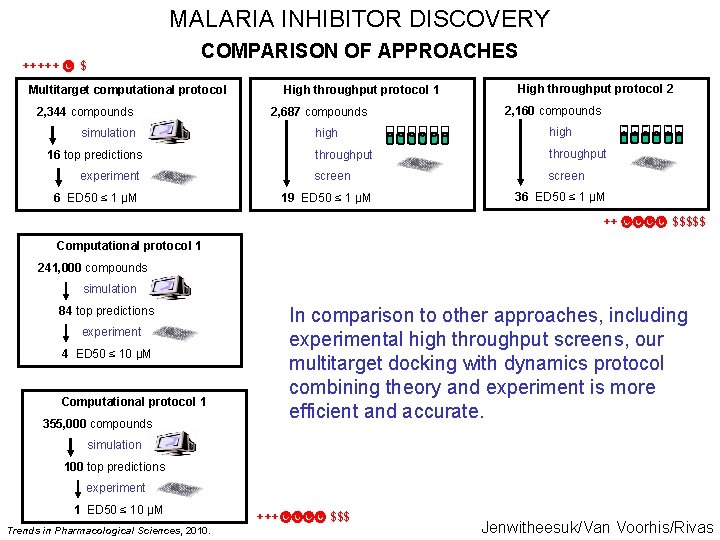
MALARIA INHIBITOR DISCOVERY +++++ COMPARISON OF APPROACHES $ Multitarget computational protocol 2, 344 compounds High throughput protocol 1 2, 687 compounds simulation 16 top predictions experiment 6 ED 50 ≤ 1 μM High throughput protocol 2 2, 160 compounds high throughput screen 19 ED 50 ≤ 1 μM 36 ED 50 ≤ 1 μM ++ $$$$$ Computational protocol 1 241, 000 compounds simulation In comparison to other approaches, including experimental high throughput screens, our multitarget docking with dynamics protocol combining theory and experiment is more efficient and accurate. 84 top predictions experiment 4 ED 50 ≤ 10 μM Computational protocol 1 355, 000 compounds simulation 100 top predictions experiment 1 ED 50 ≤ 10 μM Trends in Pharmacological Sciences, 2010. +++ $$$ Jenwitheesuk/Van Voorhis/Rivas

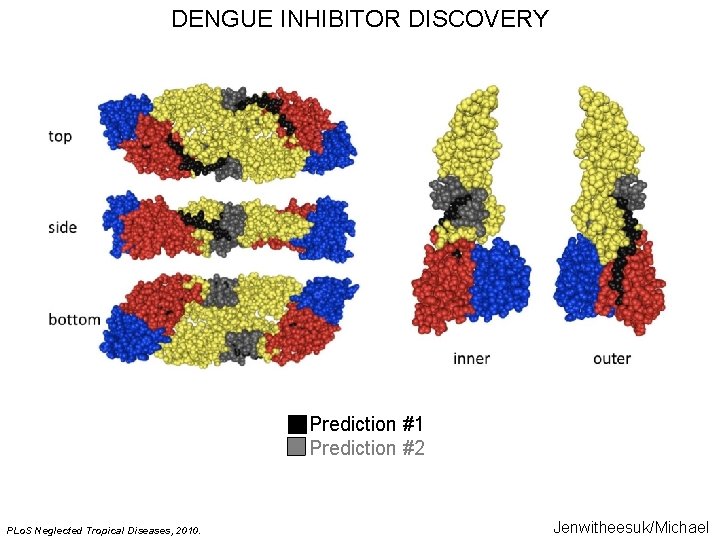
DENGUE INHIBITOR DISCOVERY Prediction #1 Prediction #2 PLo. S Neglected Tropical Diseases, 2010. Jenwitheesuk/Michael

WHY WILL IT WORK Fragment based docking with dynamics: dynamics improves accuracy; fragmentation exploits redundancy in existing drugs; most accurate docking protocol out there. Use of existing drugs: exploits all the knowledge from Pharma. Multitargeting: multiple low Kd can work synergistically; screening for targets and antitargets simultaneously. Knowledge based: potential from known structures, will have a big matrix relating drugs, targets, PK, ADME, solubility, bioavailability, toxicity, etc. ; rich dataset for combining our biophysics based methods with machine learning tools in an iterative manner. Known drugs Targets with known structure docking score, Kd, PK, ADME, absorption, bioavailability, toxicity

BROADER IMPACT Multiple drugs can be combined to produce therapeutic effect and overcome disease resistance. Good for any condition where one or more viable targets exist. Harnesses the power of all the drug discovery done thusfar; new paradigm for fast track FDA approval Translational approach goes from providing atomic mechanistic detail to measuring clinical efficacy in one shot. Protocol can be used to design novel drugs also.

SUITABILITY FOR THE PIONEER AWARD Not good for Pharma because of reuse of existing drugs (most profit in novel compounds) Not good for Pharma because of focus on third world/neglected diseases. Not good for Pharma because of nonfocus on single target model they love. Marked departure from my protein structure prediction work, but now applied research from basic protein folding to producing therapeutics in a clinic. Funding will help focus work on drug discovery which until now has been done on a shoestring.

CONCLUSION High risk endeavour is successful if one or more diseases currently without an effective treatment can be treated completely.
 Grande barrio central
Grande barrio central Nature reviews drug discovery
Nature reviews drug discovery Drug discovery process
Drug discovery process Novel clinical drug trial design
Novel clinical drug trial design Challenges of novel drug delivery system
Challenges of novel drug delivery system Methods of adulteration of crude drugs
Methods of adulteration of crude drugs Shotgun sequencing
Shotgun sequencing Blowback phenomenon wound
Blowback phenomenon wound Why do hunters pattern their shotguns
Why do hunters pattern their shotguns Pyrosequencing animation
Pyrosequencing animation Pierce electron gun
Pierce electron gun The power i offense
The power i offense Hierarchical shotgun sequencing vs whole genome
Hierarchical shotgun sequencing vs whole genome Gun barrel markings
Gun barrel markings Remington 870 shotgun nomenclature
Remington 870 shotgun nomenclature Batch processing in hci
Batch processing in hci R programming language paradigms
R programming language paradigms Message passing paradigm in distributed computing
Message passing paradigm in distributed computing 4 paradigms of cognitive psychology
4 paradigms of cognitive psychology