Normalization Getting the numbers comparable DNA Microarray Bioinformatics

Normalization Getting the numbers comparable DNA Microarray Bioinformatics - #27612
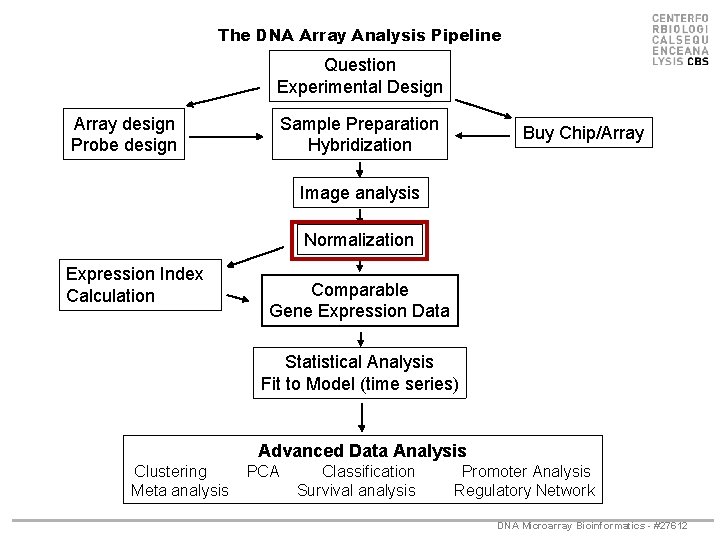
The DNA Array Analysis Pipeline Question Experimental Design Array design Probe design Sample Preparation Hybridization Buy Chip/Array Image analysis Normalization Expression Index Calculation Comparable Gene Expression Data Statistical Analysis Fit to Model (time series) Advanced Data Analysis Clustering Meta analysis PCA Classification Survival analysis Promoter Analysis Regulatory Network DNA Microarray Bioinformatics - #27612
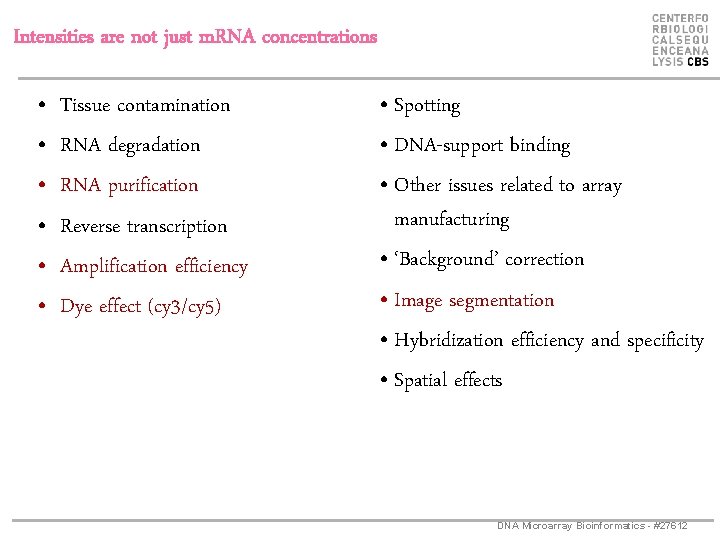
Intensities are not just m. RNA concentrations • • • Tissue contamination RNA degradation RNA purification Reverse transcription Amplification efficiency Dye effect (cy 3/cy 5) • Spotting • DNA-support binding • Other issues related to array manufacturing • ‘Background’ correction • Image segmentation • Hybridization efficiency and specificity • Spatial effects DNA Microarray Bioinformatics - #27612
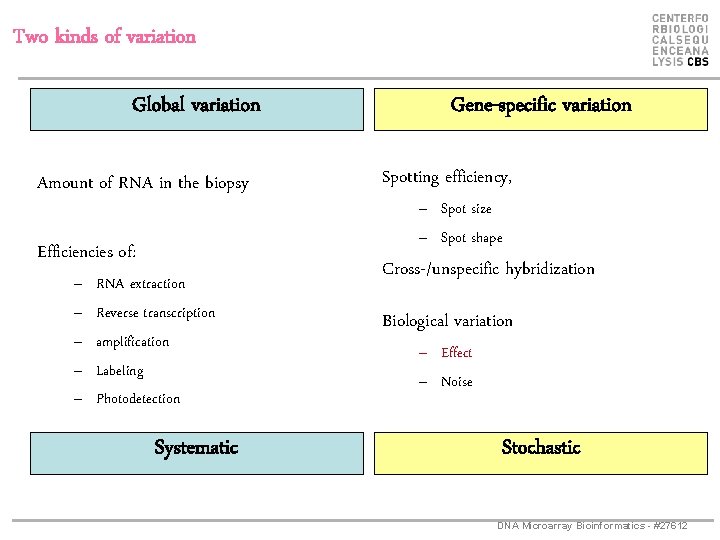
Two kinds of variation Global variation Amount of RNA in the biopsy Efficiencies of: – – – RNA extraction Reverse transcription amplification Labeling Photodetection Systematic Gene-specific variation Spotting efficiency, – Spot size – Spot shape Cross-/unspecific hybridization Biological variation – Effect – Noise Stochastic DNA Microarray Bioinformatics - #27612

Stochastic noise we use statistics to deal with PCA Plot of 34 patients, 8973 dimensions (genes) reduced to 2 DNA Microarray Bioinformatics - #27612

. . . like we will see later PCA for 100 most significant genes reduced to 2 dimensions DNA Microarray Bioinformatics - #27612
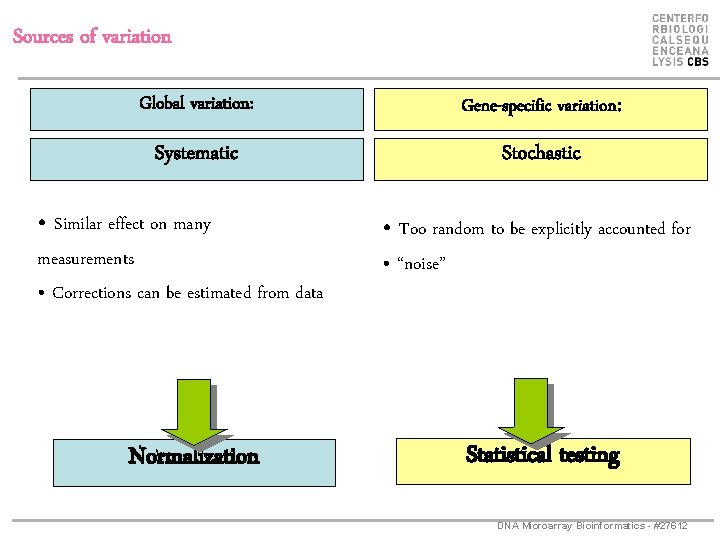
Sources of variation Global variation: Gene-specific variation: Systematic Stochastic • Similar effect on many measurements • Corrections can be estimated from data Normalization • Too random to be explicitly accounted for • “noise” Statistical testing DNA Microarray Bioinformatics - #27612
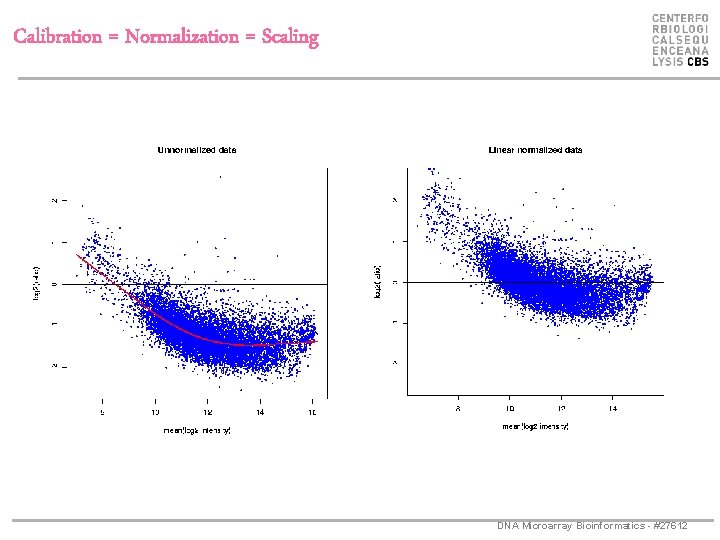
Calibration = Normalization = Scaling DNA Microarray Bioinformatics - #27612
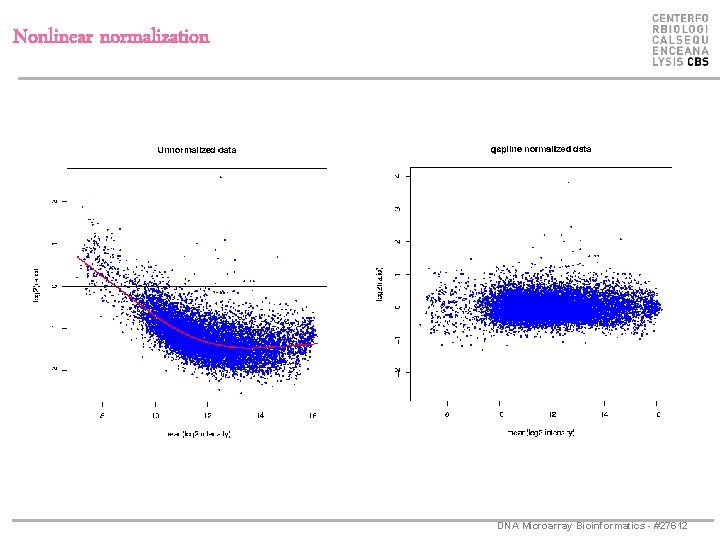
Nonlinear normalization DNA Microarray Bioinformatics - #27612
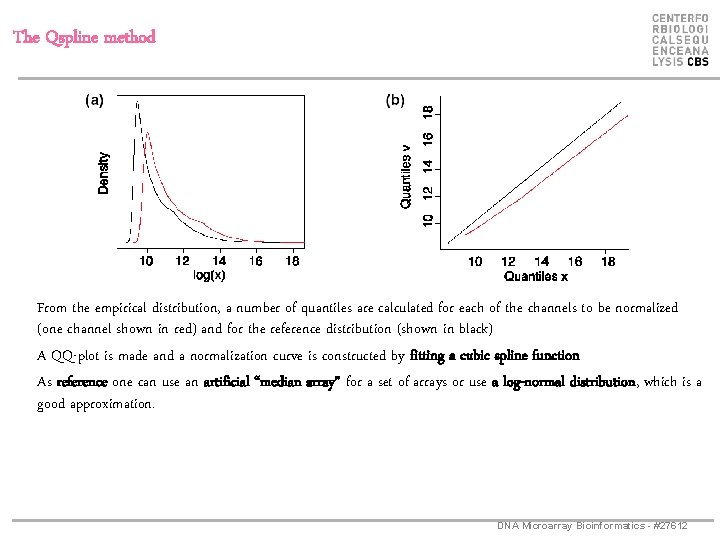
The Qspline method From the empirical distribution, a number of quantiles are calculated for each of the channels to be normalized (one channel shown in red) and for the reference distribution (shown in black) A QQ-plot is made and a normalization curve is constructed by fitting a cubic spline function As reference one can use an artificial “median array” for a set of arrays or use a log-normal distribution, which is a good approximation. DNA Microarray Bioinformatics - #27612
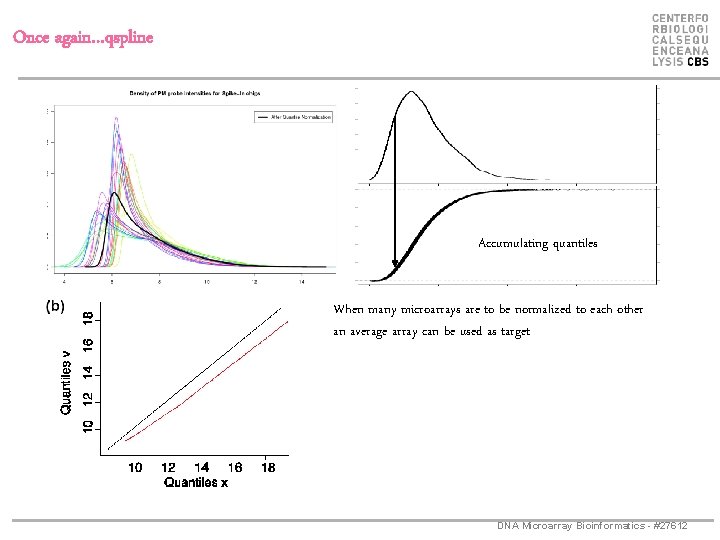
Once again…qspline Accumulating quantiles When many microarrays are to be normalized to each other an average array can be used as target DNA Microarray Bioinformatics - #27612
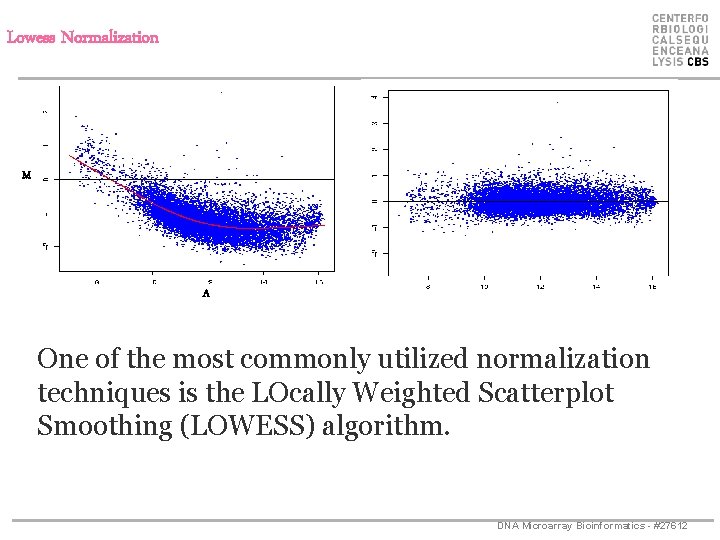
Lowess Normalization * M * * * A One of the most commonly utilized normalization techniques is the LOcally Weighted Scatterplot Smoothing (LOWESS) algorithm. DNA Microarray Bioinformatics - #27612
Invariant set normalization (Li and Wong) A invariant set of probes is used -Probes that does not change intensity rank between arrays -A piecewise linear median line is calculated -This curve is used for normalization DNA Microarray Bioinformatics - #27612
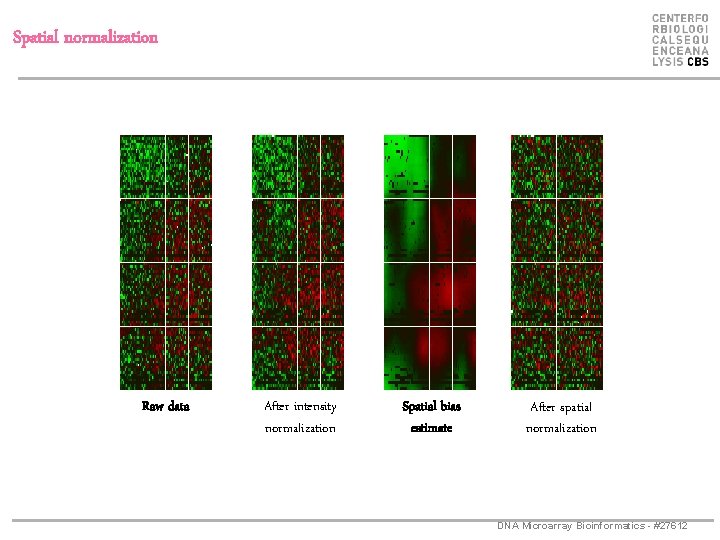
Spatial normalization Raw data After intensity normalization Spatial bias estimate After spatial normalization DNA Microarray Bioinformatics - #27612

The DNA Array Analysis Pipeline Question Experimental Design Array design Probe design Sample Preparation Hybridization Buy Chip/Array Image analysis Normalization Expression Index Calculation Comparable Gene Expression Data Statistical Analysis Fit to Model (time series) Advanced Data Analysis Clustering Meta analysis PCA Classification Survival analysis Promoter Analysis Regulatory Network DNA Microarray Bioinformatics - #27612
Expression index value Some microarrays have multiple probes addressing the expression of the same gene – Affymetrix chips have 11 -20 probe pairs pr. Gene - Perfect Match (PM) - Mis. Match (MM) PM: MM: CGATCAATTGCACTATGTCATTTCT CGATCAATTGCAGTATGTCATTTCT DNA Microarray Bioinformatics - #27612
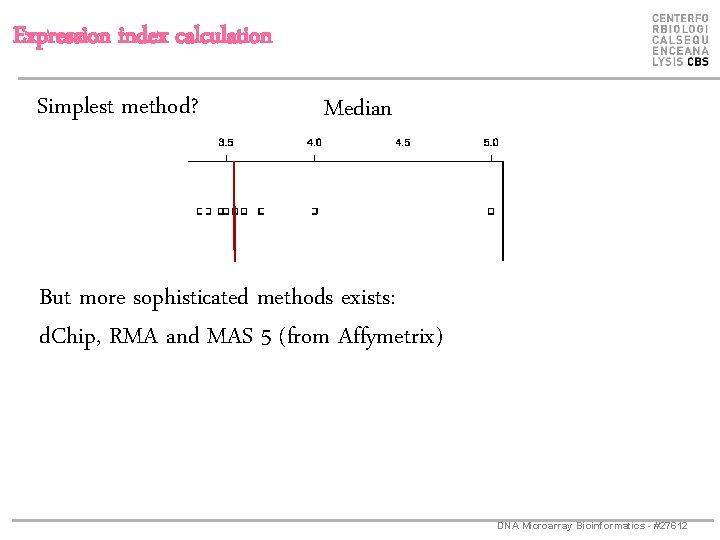
Expression index calculation Simplest method? Median But more sophisticated methods exists: d. Chip, RMA and MAS 5 (from Affymetrix) DNA Microarray Bioinformatics - #27612
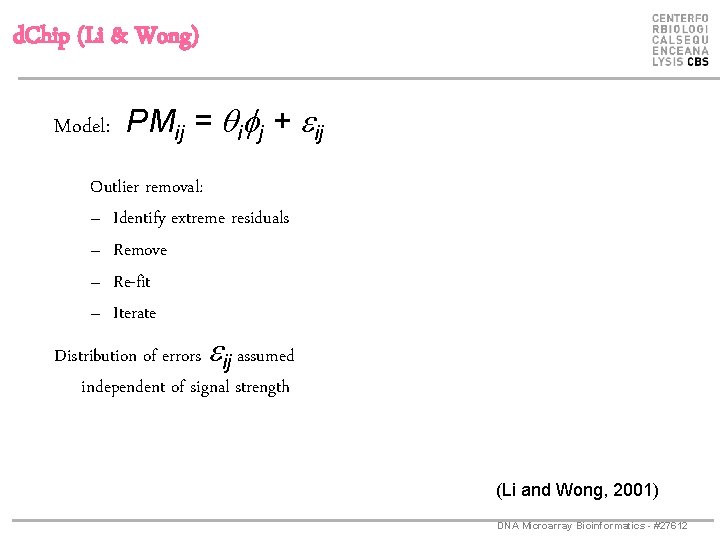
d. Chip (Li & Wong) Model: PMij = qifj + eij Outlier removal: – Identify extreme residuals – Remove – Re-fit – Iterate Distribution of errors eij assumed independent of signal strength (Li and Wong, 2001) DNA Microarray Bioinformatics - #27612
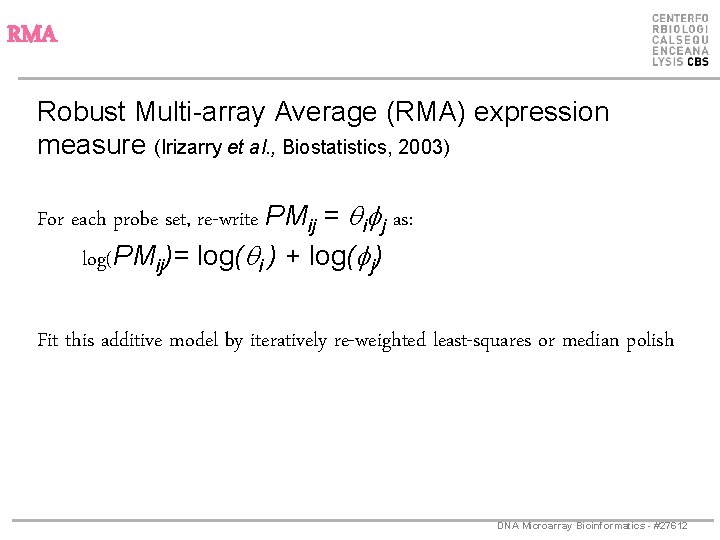
RMA Robust Multi-array Average (RMA) expression measure (Irizarry et al. , Biostatistics, 2003) For each probe set, re-write PMij = qifj as: log(PMij)= log(qi ) + log(fj) Fit this additive model by iteratively re-weighted least-squares or median polish DNA Microarray Bioinformatics - #27612

MAS. 5 Micro. Array Suite version 5 uses MM* is an adjusted MM that is never bigger than PM Tukey biweight is a robust average procedure with weights and outlier rejection DNA Microarray Bioinformatics - #27612
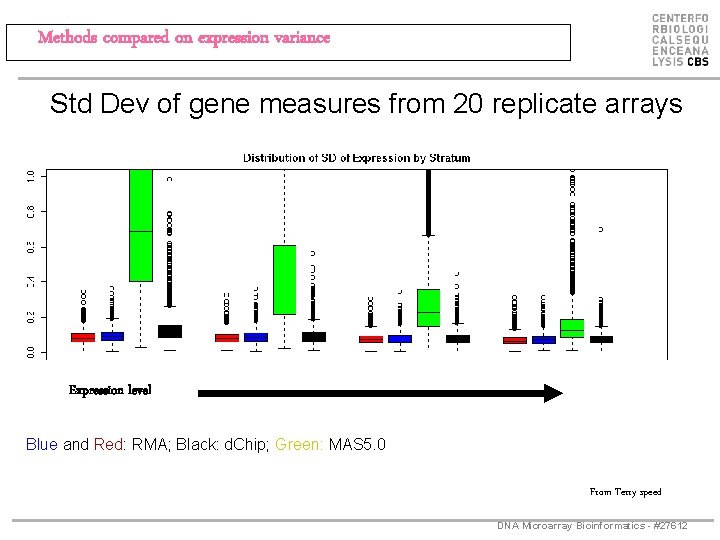
Methods compared on expression variance Std Dev of gene measures from 20 replicate arrays Expression level Blue and Red: RMA; Black: d. Chip; Green: MAS 5. 0 From Terry speed DNA Microarray Bioinformatics - #27612

Robustness MAS 5. 0 Log fold change estimate from 20 ug c. RNA MAS 5. 0 Log fold change estimate from 1. 25 ug c. RNA (Irizarry et al. , Biostatistics, 2003) DNA Microarray Bioinformatics - #27612

Robustness d. Chip Log fold change estimate from 20 ug c. RNA d. Chip Log fold change estimate from 1. 25 ug c. RNA (Irizarry et al. , Biostatistics, 2003) DNA Microarray Bioinformatics - #27612
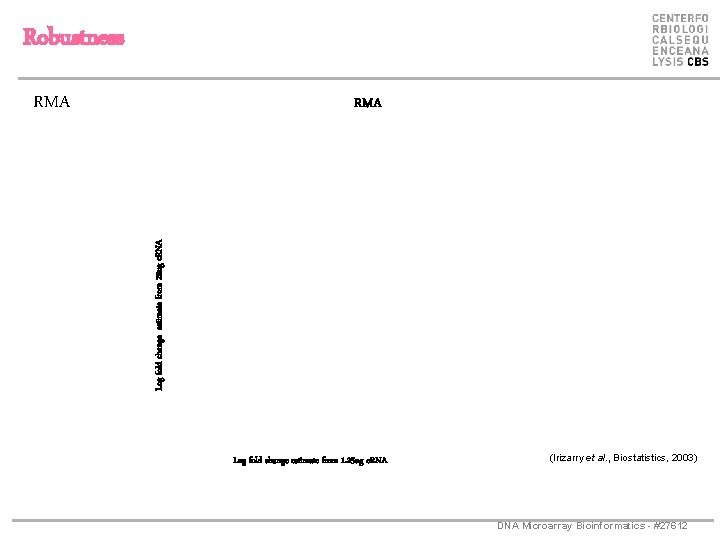
Robustness RMA Log fold change estimate from 20 ug c. RNA RMA Log fold change estimate from 1. 25 ug c. RNA (Irizarry et al. , Biostatistics, 2003) DNA Microarray Bioinformatics - #27612

All of this is implemented in… R In the Bio. Conductor packages ‘affy’ (Gautier et al. , 2003). DNA Microarray Bioinformatics - #27612

References Li and Wong, (2001). Model-based analysis of oligonucleotide arrays: Model validation, design issues and standard error application. Genome Biology 2: 1– 11. Irizarry, Bolstad, Collin, Cope, Hobbs and Speed, (2003) Summaries of Affymetrix Gene. Chip probe level data. Nucleic Acids Research 31(4): e 15. ) Affymetrix Microarray Suite User Guide. Affymetrix, Santa Clara, CA, version 5 edition, 2001. Gautier, Cope, Bolstad, and Irizarry, (2003). affy - an r package for the analysis of affymetrix genechip data at the probe level. Bioinformatics DNA Microarray Bioinformatics - #27612
- Slides: 26