Normal DNA Seq Tumor RNA Seq Alignment BWA
Normal DNA Seq Tumor RNA Seq Alignment (BWA) Alignment (Tophat 2) Expression estimation (cufflinks) Variant calling (gatk) Expression levels in FPKM Variant list In-silico HLA Haplotyping (Opti. Type and seq 2 HLA) Variant annotation (VEP - ensembl) Annotated variant list Filters: - Median IC 50 < 500 n. M - Median fold change > 2 Epitope prediction (pvacseq) Epitope candidates

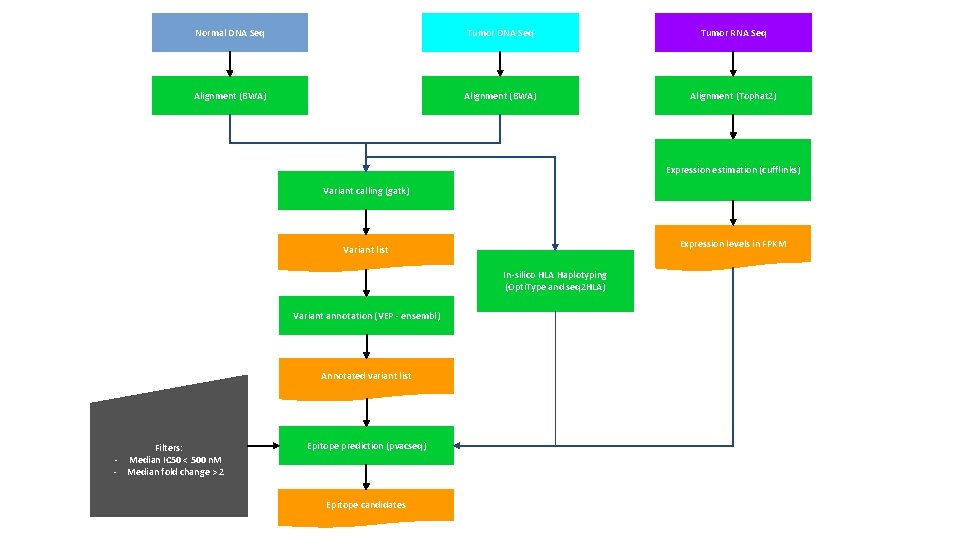
Normal DNA Seq Tumor RNA Seq Alignment (BWA) Alignment (Tophat 2) Expression estimation (cufflinks) Variant calling (gatk) Expression levels in FPKM Variant list In-silico HLA Haplotyping (Opti. Type and seq 2 HLA) Variant annotation (VEP ensembl) Annotated variant list Filters: - Median IC 50 < 500 n. M - Median fold change > 2 Epitope prediction (pvacseq) Epitope candidates Fig. X Overview of the pipeline for identifying and prioritizing cancer neoantigens: This figure illustrates the methodology implemented so far

Normal DNA Seq Tumor RNA Seq Alignment (BWA) Alignment (Tophat 2) Expression estimation (cufflinks) Variant calling (gatk) Variant list Variant annotation (VEP - ensembl) In-silico HLA Haplotyping (Opti. Type) Expression levels in FPKM Annotated variant list Filters: Median IC 50 < 100 n. M - Median fold change > 10 - Epitope prediction (pvacseq) Epitope candidates Step performed Step missing Fig. X Overview of the pipeline for identifying and prioritizing cancer neoantigens: This figure illustrates the methodology implemented so far
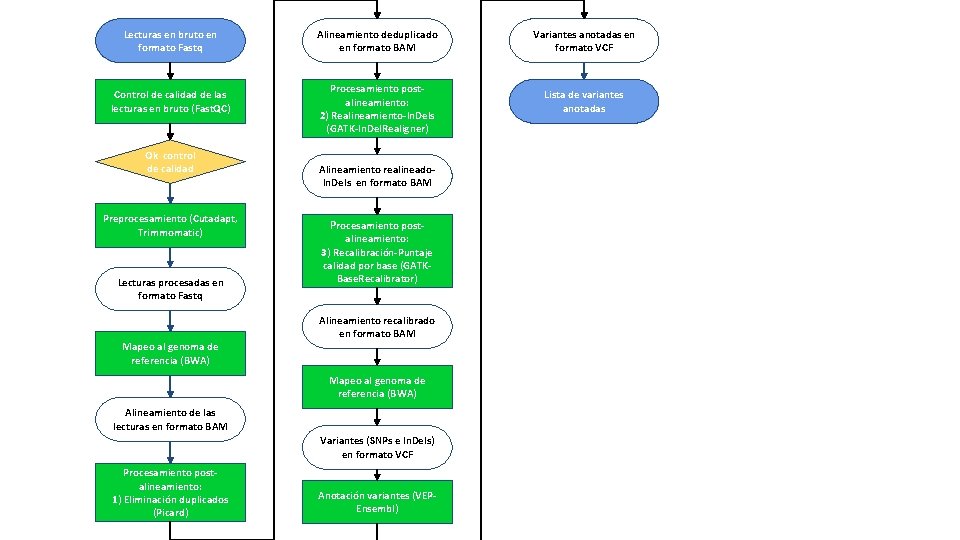
Lecturas en bruto en formato Fastq Control de calidad de las lecturas en bruto (Fast. QC) Ok control de calidad Preprocesamiento (Cutadapt, Trimmomatic) Lecturas procesadas en formato Fastq Alineamiento deduplicado en formato BAM Procesamiento postalineamiento: 2) Realineamiento-In. Dels (GATK-In. Del. Realigner) Alineamiento realineado. In. Dels en formato BAM Procesamiento post- alineamiento: 3) Recalibración-Puntaje calidad por base (GATKBase. Recalibrator) Alineamiento recalibrado en formato BAM Mapeo al genoma de referencia (BWA) Alineamiento de las lecturas en formato BAM Variantes (SNPs e In. Dels) en formato VCF Procesamiento postalineamiento: 1) Eliminación duplicados (Picard) Anotación variantes (VEPEnsembl) Variantes anotadas en formato VCF Lista de variantes anotadas
- Slides: 4