NIDA Genetics An Update Jonathan Pollock Ph D













- Slides: 45

NIDA Genetics: An Update Jonathan Pollock, Ph. D Branch Chief, Genetics and Molecular Neurobiology Research Branch DBNBR And Joni L Rutter, Ph. D Associate Director for Population and Applied Genetics DBNBR NIDA NATIONAL INSTITUTE ON DRUG ABUSE

Individual Differences in Vulnerability to Addiction n Not everyone who takes drugs becomes addicted n “has characteristics of chronic disease” Piazza Science 13 August 2004 There are individual differences n Chronic exposure to drugs combined with a vulnerability phenotype leads to addiction n Genetics contributes to some of these individual differences n

Drug Addiction Is Influenced by Interactions of Genes and Environment Twin studies consistently show that there is a heritable component to drug abuse and addiction. Now we are able to examine genetic variants, or single nucleotide polymorphisms that contribute to addiction vulnerability

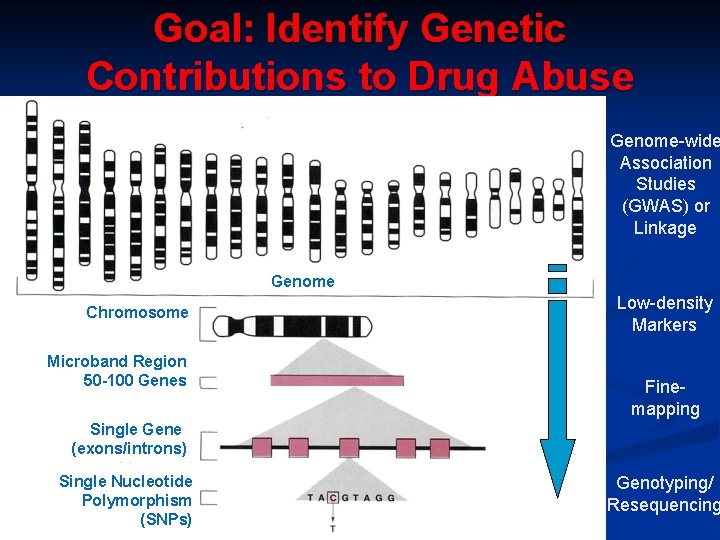
Goal: Identify Genetic Contributions to Drug Abuse Genome-wide Association Studies (GWAS) or Linkage Genome Chromosome Microband Region 50 -100 Genes Low-density Markers Finemapping Single Gene (exons/introns) Single Nucleotide Polymorphism (SNPs) Genotyping/ Resequencing

NIDA Genetics: Identify Genetic Contributions to Drug Abuse n 2 Main Programmatic Themes n Trans-NIH initiatives n Leverage ongoing programs n Roadmap, GAIN, GEI, PGRN, etc n Trans-NIDA programs n Coordination among divisions n Multidisciplinary/Interdisciplinary n Data

Human Genetics Trans-NIH Involvement n Roadmap 1. 0 n National Centers for Biomedical Computing (NCBC) - NIDA Lead, Karen Skinner n i 2 b 2 n n n (informatics for integrating biology and the bedside) Zak Kohane, PI--evaluate smoking data in medical records forge collaborations on genetics and bioinformatics of smoking Roadmap 1. 5 – Epigenomics n Nora Volkow, John Satterlee, Christine Colvis, Genevieve de. Almeida-Morris, Jonathan Pollock, David Shurtleff, Joni Rutter http: //nihroadmap. nih. gov/epigenomics/

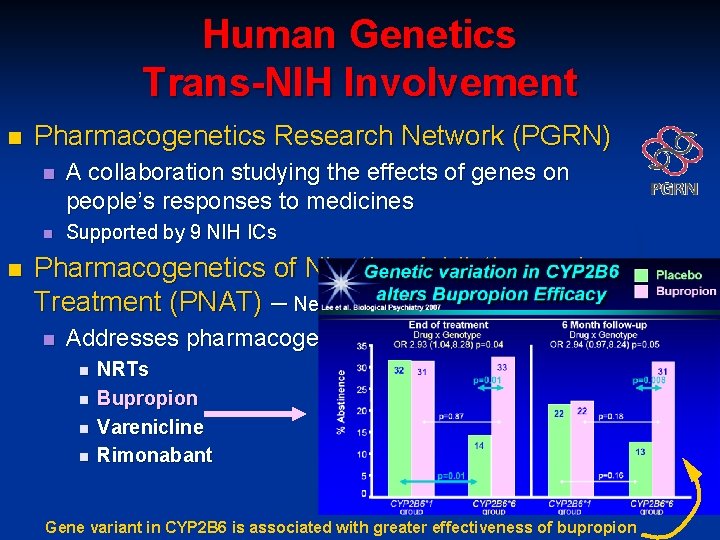
Human Genetics Trans-NIH Involvement n n Pharmacogenetics Research Network (PGRN) n A collaboration studying the effects of genes on people’s responses to medicines n Supported by 9 NIH ICs Pharmacogenetics of Nicotine Addiction and Treatment (PNAT) – Neal Benowitz, PI n Addresses pharmacogenetics of nicotine addiction n n NRTs Bupropion Varenicline Rimonabant Gene variant in CYP 2 B 6 is associated with greater effectiveness of bupropion

Human Genetics Trans-NIH Involvement n Genetic Association Information Network (GAIN) n n n Public-private partnership led by Foundation for NIH Genome-wide association study policy development for NIH-- Jonathan Pollock Genes, Environment and Health Initiative (GEI) n Genetics Program- Pipeline for analyzing genetic variation in groups of patients with specific illnesses n n Genetics of Addiction – Laura Bierut, PI Exposure Biology Program- Measuring environmental exposures n NIDA RFA: Technologies measuring exposure to psychosocial stress and addictive substances--Kay Wanke, Kevin Conway

NIDA Genetics: Identify Genetic Contributions to Drug Abuse n 2 Main Programmatic Themes n Trans-NIH initiatives n Leverage ongoing programs n Roadmap, GAIN, GEI, PGRN, etc n Trans-NIDA programs n Coordination among divisions n Multidisciplinary/Interdisciplinary n Data

NIDA Genetics: A Coordinated Effort Model Organisms NIDA Genetics Workgroup Members NGCC members Christine Colvis Jim Glass Hal Gordon Mark Green Jag Khalsa Diane Lawrence Mary Ellen Michel Ivan Montoya Jonathan Pollock, Chair John Satterlee David Shurtleff Karen Skinner Mark Swieter Yonette Thomas George Uhl (IRP) Susan Volman Da-Yu Wu Scientific Goals & Services § Develop avenues of research through FOAs § Sponsor seminars to inform NIDA of new areas of genetics research § Develop staff knowledge in areas of genetics research Human NIDA Genetic Coordinating Committee (NGCC) Members Beth Babecki Kevin Conway Ahmed Elkashef Steve Grant Rita Liu Raul Mandler Cindy Miner JJ Pan Amrat Patel Jonathan Pollock Joni Rutter, Chair Larry Stanford Kay Wanke Naimah Weinberg Scientific Goals & Services §Written ARA, supplement, and funding recommendations to the Director §Recommend genetic applications assignment §Provide standards for data collection, data sharing, and informed consent §Evaluate human genetics program §Serve as the NGC Steering Committee

Human Genetics Trans-NIDA Genetics n NIDA Genetics Consortium (NGC) n n NIDA Division Reps (+1 NCI rep) >20 PIs; 24 studies; 1 contract n n ~30, 000 samples in Repository n n http: //zork. wustl. edu/nida/ DNA and clinical information (DSMIIIR or IV) Nicotine, Opioids, Cocaine, Polysubstance Data to be publicly available through controlled access NIDA Phenotyping Consortium (NPC) n n DESPR-led initiative (Kevin Conway); 5 PIs Produce precise and specific phenotypes for drug abuse Addiction “Bioseverity” Develop a research instrument measuring addiction severity Validated with biological processes (ex. Genetic, imaging, etc) Michael Vanyukov (NGC), Gary Swan (NGC), Michael Neale (NPC) Kevin Conway (DESPR), Janet Levy (DESPR), Joni Rutter (DBNBR)

Human Genetics Trans-NIDA Genetics n Collaboration between NGC, CTN, DPMCDA n n “START” Study – Starting Treatment on Agonist Replacement Therapies –added pharmacogenetics Targeting ~600 opioid dependent persons Randomized, open-labeled CTN multi-site design to either buprenorphine or methadone Primary outcomes are: n n Liver toxicity (parent trial) Individual genetic variation in treatment effectiveness (Pharmacogenetics) n Wade Berrettini and Lindsay De. Vane

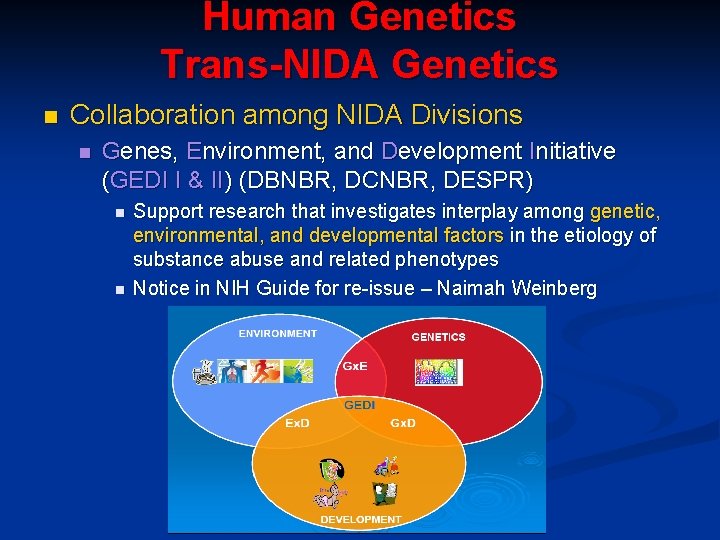
Human Genetics Trans-NIDA Genetics n Collaboration among NIDA Divisions n Genes, Environment, and Development Initiative (GEDI I & II) (DBNBR, DCNBR, DESPR) n n Support research that investigates interplay among genetic, environmental, and developmental factors in the etiology of substance abuse and related phenotypes Notice in NIH Guide for re-issue – Naimah Weinberg

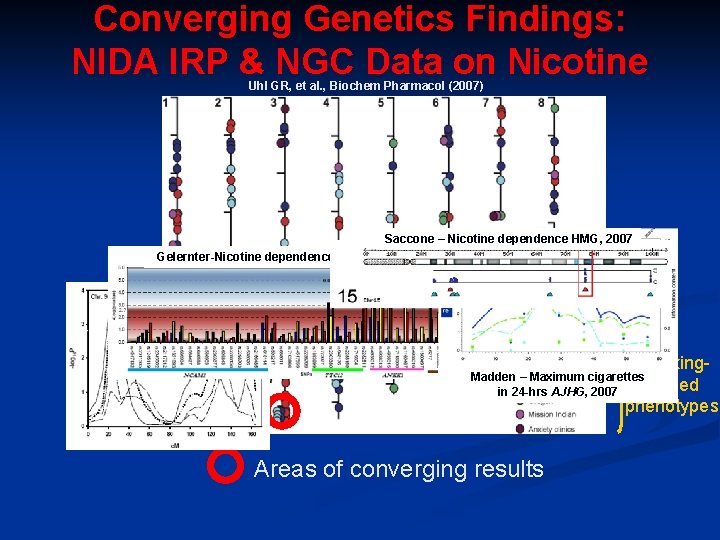
Converging Genetics Findings: NIDA IRP & NGC Data on Nicotine Uhl GR, et al. , Biochem Pharmacol (2007) Saccone – Nicotine dependence HMG, 2007 Gelernter-Nicotine dependence HMG, 2006 Li - Nicotine dependence AJHG, 2006 Smoking- Madden – Maximum cigarettes related in 24 -hrs AJHG, 2007 phenotypes Areas of converging results

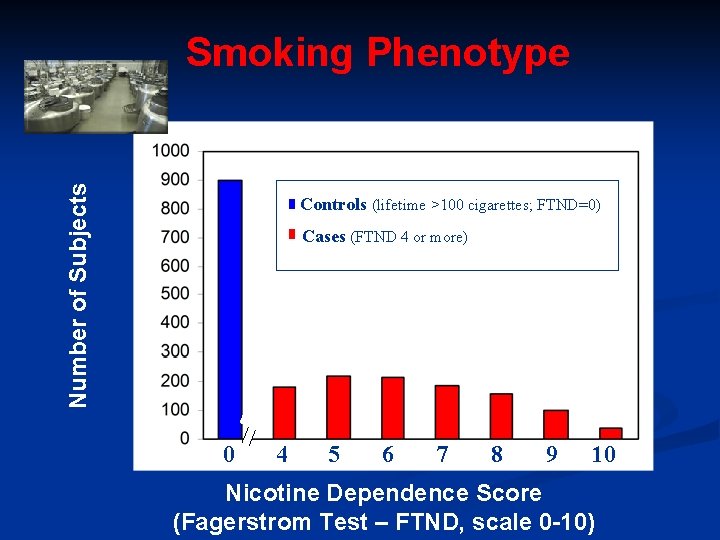
Number of Subjects Smoking Phenotype Controls (lifetime >100 cigarettes; FTND=0) Cases (FTND 4 or more) 0 4 5 6 7 8 9 10 Nicotine Dependence Score (Fagerstrom Test – FTND, scale 0 -10)

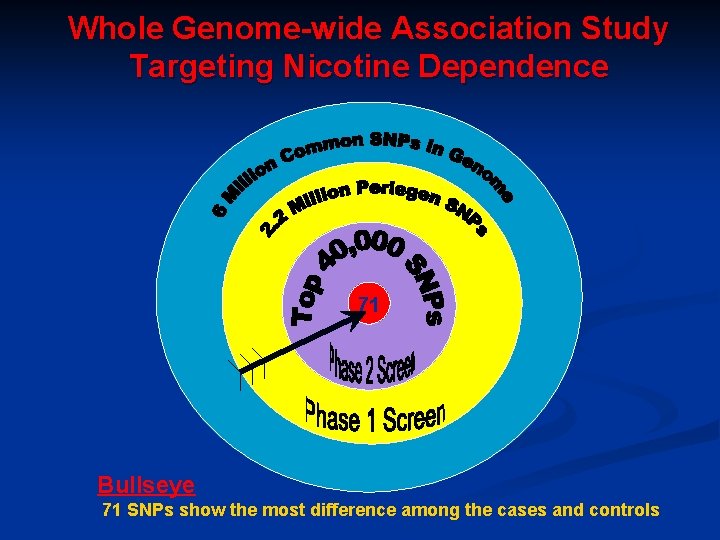
Whole Genome-wide Association Study Targeting Nicotine Dependence 71 Bullseye 71 SNPs show the most difference among the cases and controls

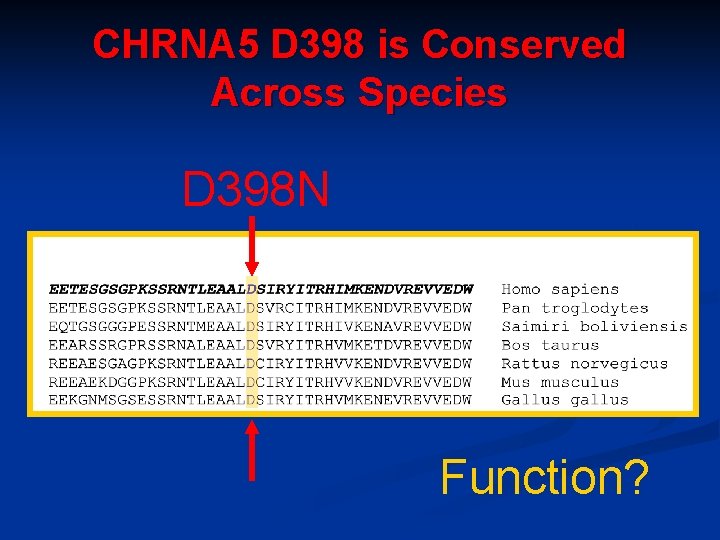
Several SNPS Found to be Different Between Cases and Control 71 SNPS different between case and control n Many were not the “usual suspects” n Several nicotinic receptors implicated : n n a 3 nicotinic receptor n β 3 nicotinic receptor n a 5 nicotinic receptor SNP is highly associated with nicotine dependence **CHRNA 5 non-synonymous SNP D 398 N Individuals with this SNP have 2 -fold increase risk of developing nicotine dependence OR=1. 9 (1. 4 -2. 6) Saccone et al. HMG 2007 Bierut et al. HMG 2007

CHRNA 5 D 398 is Conserved Across Species D 398 N Function?

The human CHRNA 5 D 398 N polymorphism alters nicotinic receptor function in vitro Common allele Variant allele P < 0. 01 ** Courtesy of Jerry Stitzel, Ph. D

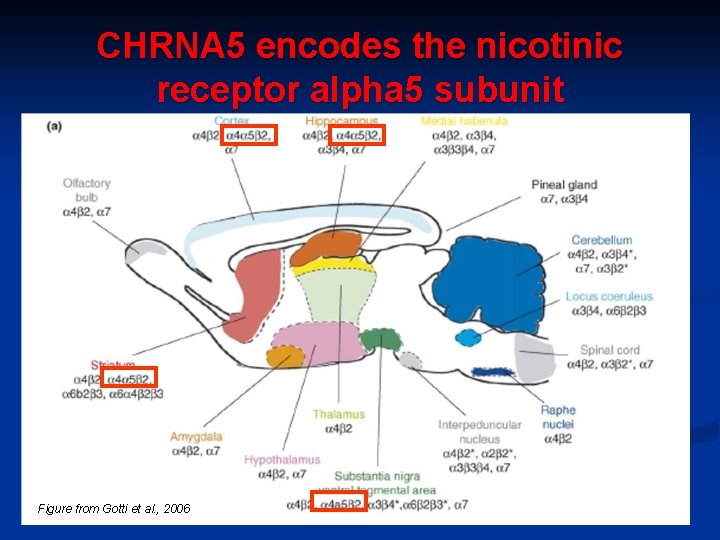
CHRNA 5 encodes the nicotinic receptor alpha 5 subunit Figure from Gotti et al. , 2006

Summary n Trans-NIH involvement n n Roadmap, GAIN, GEI, PGRN, others NIDA staff play leadership roles NIDA PIs positioned to take advantage of programs NIDA Genetics n n Divisional coordination, programmatic synergy Phenotype, environment, development, family, clinical, animal models Results are converging Functional information Mechanism Inform treatment, prevention, intervention approaches Molecular Genetics of Drug Addiction and Related Co-Morbidities (R 01) PA-07 -073 (R 01) - November 20, 2006 Functional Genetics and Genomics of Drug Addiction PA-07 -166 (R 01), PA-07 -167 (R 21), PA-07 -168 (R 03) - December 14, 2006 Genetic Epidemiology of Substance Use Disorders PA-07 -413 (R 01), PA-07 -415 (R 21), PA-07 -414 (R 03) - July 27, 2007

Drug Abuse Genetics Environment/ Epidemiology Genes Family/ Clinical Model organisms Development

Genetic model organisms, such as the mouse, can provide clues to the genetic basis of complex diseases

Three Different Methods to Identify Gene Function in Mice n Haplotype Associated Mapping of Natural Variation in Inbred Strains n Selective Breeding n Induced Mutations, e. g. Knockouts

Three Different Methods to Identify Gene Function in Mice n Haplotype Associated Mapping of Natural Variation in Inbred Strains n Selective Breeding n Induced Mutations, e. g. Knockouts

Power of Inbred Strains n Inbred strains are a way to determine environmental effects through development across defined genotypes under controlled conditions n Confidence in the underlying genotype is strengthened when phenotypes map to homologous chromosomal loci in mice and humans

C 3 H/He. J C 57 L/J I/Ln. J C 57 BL/6 J LP/J CE/J A/J DBA/2 J C 57 BR/cd. J

Haplotype Associated Mapping of Natural Variation in Inbred Strains n Many mouse genomes have been sequenced e. g. C 57 BL 6/J, AJ, 129 n Genetic variation in over 15 strains of mice have been identified n Use variation in gene expression profiling as a “signature” for a trait

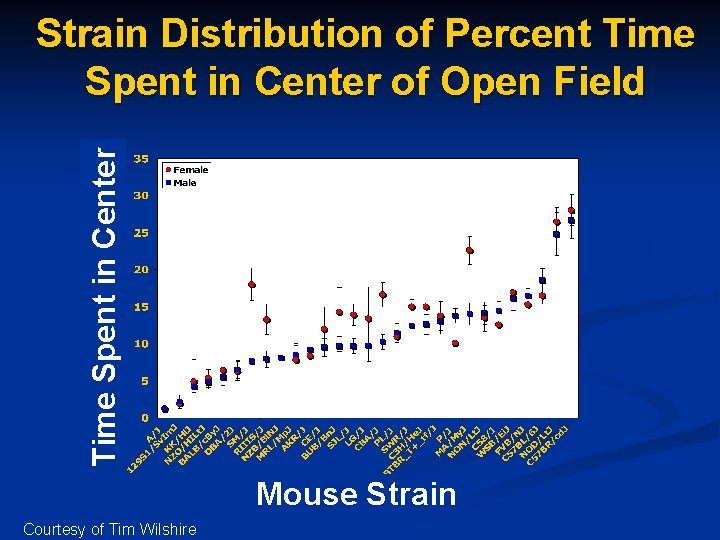
Time Spent in Center Strain Distribution of Percent Time Spent in Center of Open Field Mouse Strain Courtesy of Tim Wilshire

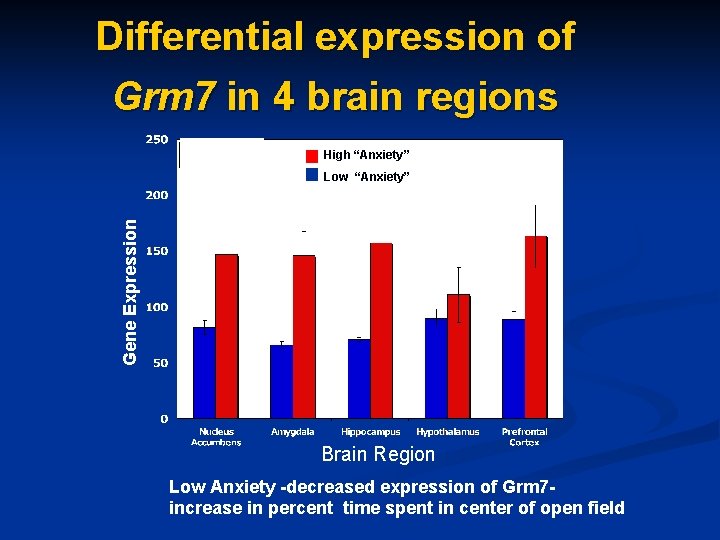
Differential expression of Grm 7 in 4 brain regions High “Anxiety” Gene Expression Low “Anxiety” Brain Region Low Anxiety -decreased expression of Grm 7 increase in percent time spent in center of open field

Three Different Methods to Identify Gene Function in Mice n Haplotype Associated Mapping of Natural Variation in Inbred Strains n Selective Breeding n Induced Mutations, e. g. Knockouts

Mapping Genes Using Selective Breeding n Takes advantage of the mouse genome and gene variants (SNPs) among different mouse strains n NIH proposed project will increase genetic diversity in inbred recombinant mice making it possible to map any trait in the mouse (i. e. , The “Collaborative Cross”)

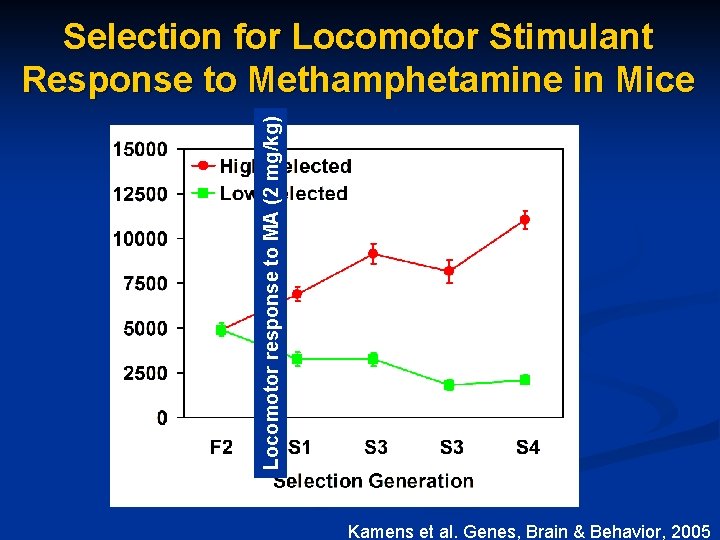
Locomotor response to MA (2 mg/kg) Selection for Locomotor Stimulant Response to Methamphetamine in Mice Kamens et al. Genes, Brain & Behavior, 2005

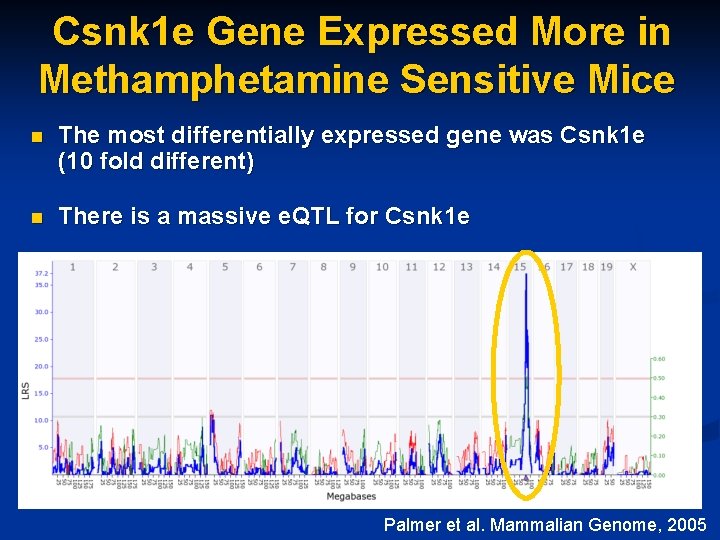
Csnk 1 e Gene Expressed More in Methamphetamine Sensitive Mice n The most differentially expressed gene was Csnk 1 e (10 fold different) n There is a massive e. QTL for Csnk 1 e Palmer et al. Mammalian Genome, 2005

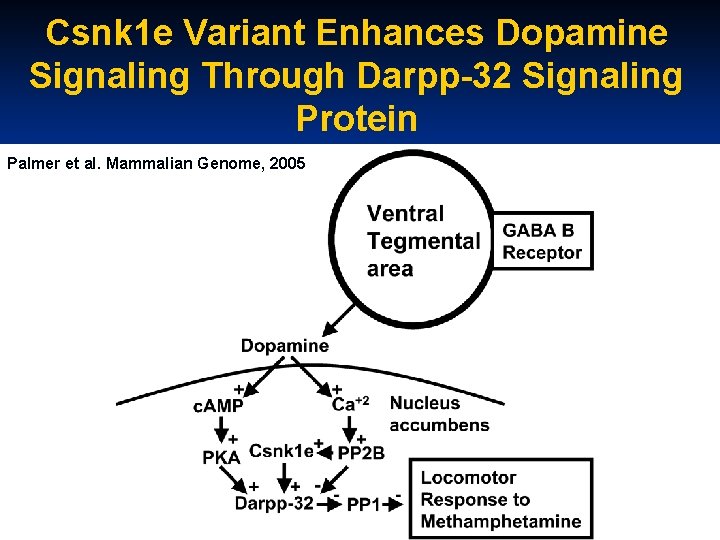
Csnk 1 e Variant Enhances Dopamine Signaling Through Darpp-32 Signaling Protein Palmer et al. Mammalian Genome, 2005

Translational Genetic Models of Methamphetamine Sensitivity n Humans vary in methamphetamine sensitivity n Differences in sensitivity are partially under genetic control in both mice and humans n Differences in initial effects of drugs linked to future use n An predictor or intermediate phenotype to addiction in humans

Effect of CSNK 1 E Polymorphisms on Response to Methamphetamine • 3 polymorphisms in CSNK 1 E in humans – All 3 SNPs occur at high frequency • Analyzed their relationship with 3 primary outcome measures: – DEQ “Feel Drug” – ACRI “Euphoria” – POMS “Anxiety” Veenstra-Vander. Weele et al. Neuropsychopharm. , 2005

ACRI “Euphoria” is Also Significantly Influenced by SNPs in CSNK 1 E * * * Veenstra-Vander. Weele et al. Neuropsychopharmacology, 2005

Chromosomal Regions are Conserved for Drug Response in Mice and Humans Formin n LRP 2 n Alpha 6 integrin n NFI/B n NPAS 2 n NIMA-related 7 Kinase n Uhl et al, 2007 Biochemical Pharmacology

Three Different Methods to Identify Gene Function in Mice n Haplotype Associated Mapping of Natural Variation in Inbred Strains n Selective Breeding n Induced Mutations, e. g. Knockouts

NIH Knockout Mouse Project (KOMP) n KOMP is a trans-NIH project involving 18 Institutes n The NIH has committed ~ $52 M over 5 years n KOMP is cooperating with other international efforts to minimize overlap

Why are Knockouts and Transgenic Animals Useful? n Help identify essential molecules in reward pathways mediating drug abuse n Help separate cellular signaling pathways that mediate different phenomena associated with addiction (e. g. , withdrawal, tolerance)

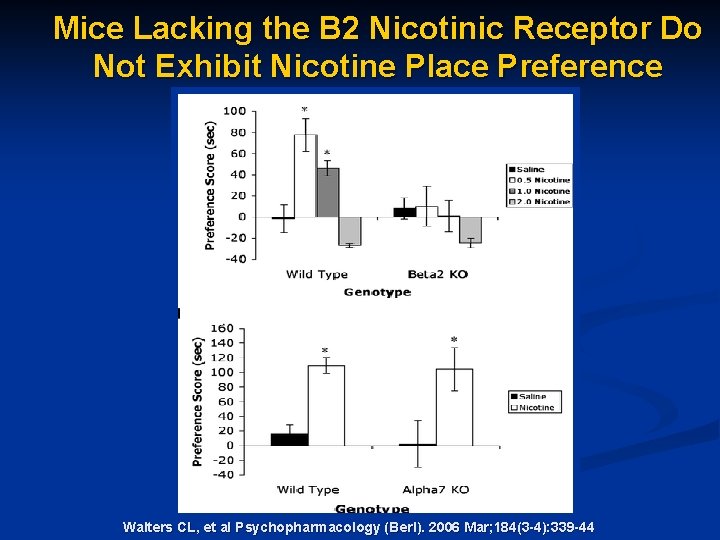
Mice Lacking the Β 2 Nicotinic Receptor Do Not Exhibit Nicotine Place Preference . Walters CL, et al Psychopharmacology (Berl). 2006 Mar; 184(3 -4): 339 -44

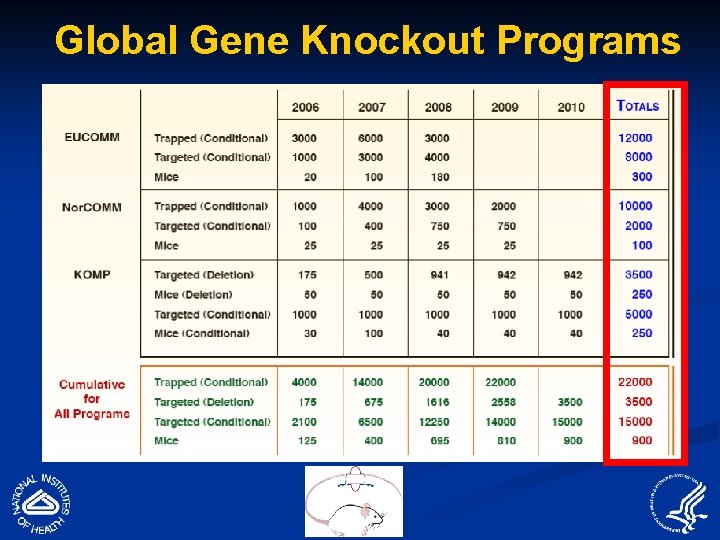
Global Gene Knockout Programs

Model Drug Abuse Organism Genetics Environment Genes knockout Epigenetics Development