NextGeneration Sequencing of Microbial Genomes and Metagenomes Christine

Next-Generation Sequencing of Microbial Genomes and Metagenomes Christine King Farncombe Metagenomics Facility Human Microbiome Journal Club July 13, 2012
Overview Next-generation sequencing � Applications � Instruments � Library prep and sequencing chemistry � Sequence quality Project overview � Microbial genomes � Microbial communities

DNA Sequencing 1 st generation � Sanger chain termination � Capillary electrophoresis 2 nd generation (NGS) � High throughput, “massively parallel” � Shorter reads � Sequencing-bysynthesis 3 rd generation � Single molecule
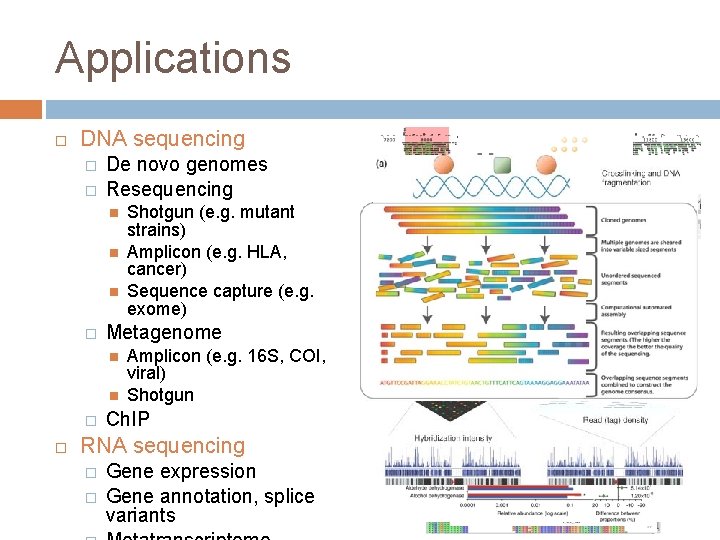
Applications DNA sequencing � � De novo genomes Resequencing � Metagenome � Shotgun (e. g. mutant strains) Amplicon (e. g. HLA, cancer) Sequence capture (e. g. exome) Amplicon (e. g. 16 S, COI, viral) Shotgun Ch. IP RNA sequencing � � Gene expression Gene annotation, splice variants

Instruments

Instruments Instrument # of read s Read length (bp) Total outp ut (Gb) Cost per base Run Time GS FLX 1 M 450 0. 5 $$$$ ++ GS FLX+ 1 M 650 0. 6 $$$$ ++ GS Jr 100 K 450 0. 05 $$$$ ++ GAIIx 640 M 2 x 150 90 $$ +++ Hi. Seq 2000 6 B 2 x 100 600 $ +++ Mi. Seq 12 M 2 x 150 2 $$ ++ Pac. Bio RS >10 K >1000 0. 01 $$$$ + Single-molecule seq, fluorophore SOLi. D 5500 xl 1. 4 B 75 + 35 155 $ +++ em. PCR, probe ligation, fluorophore Ion PGM 316 1 M >100 0. 1 $$$ + Ion PGM 318 6 M Technology em. PCR, SBS, light detection Bridge PCR, SBS, fluororphore em. PCR, SBS, p. H change >100 1 $$ +

Which instrument(s) to use? Read length vs number of reads Cost per base, per sample, per project (multiplexing? ) Accuracy Run time, wait time Application Lengt # h Reads Accura cy Instruments Considerations De novo (small) +++ ++ ++ Mi. Seq, 454, Ion Mix lengths De novo (large) +++ ++ Hi. Seq, 454, SOLi. D Mix lengths, MP Re-seq (small) ++ ++ ++ Mi. Seq, Ion Multiplex? Re-seq (large) ++ ++ Hi. Seq, SOLi. D Enrichment? RNA-seq (count) +++ + Illumina, SOLi. D, Ion Ref? Size? Rare? +
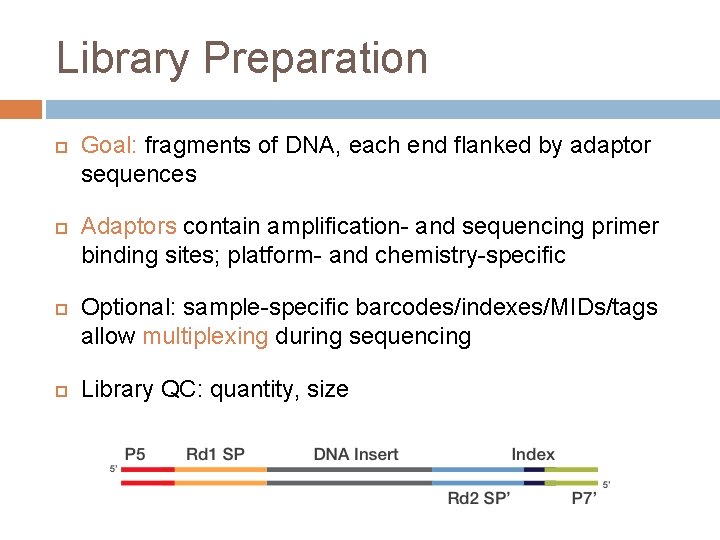
Library Preparation Goal: fragments of DNA, each end flanked by adaptor sequences Adaptors contain amplification- and sequencing primer binding sites; platform- and chemistry-specific Optional: sample-specific barcodes/indexes/MIDs/tags allow multiplexing during sequencing Library QC: quantity, size
Library Preparation Library types: � Shotgun (DNA) May begin with Ch. IP May follow with sequence capture � Mate pair (DNA) � Amplicon (DNA) � Total RNA May enrich for m. RNA (poly-A enrichment, r. RNA depletion) Convert to c. DNA (then similar to DNA protocols) � Small RNA ligations, convert to c. DNA after
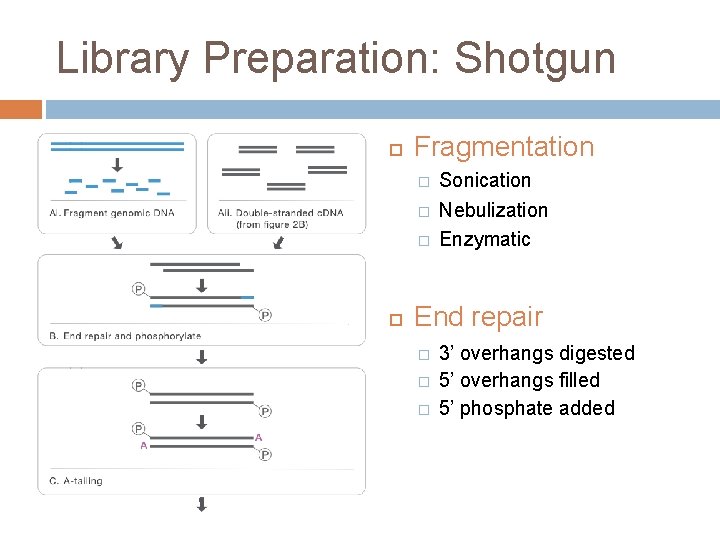
Library Preparation: Shotgun Fragmentation � � � Sonication Nebulization Enzymatic End repair � � � 3’ overhangs digested 5’ overhangs filled 5’ phosphate added
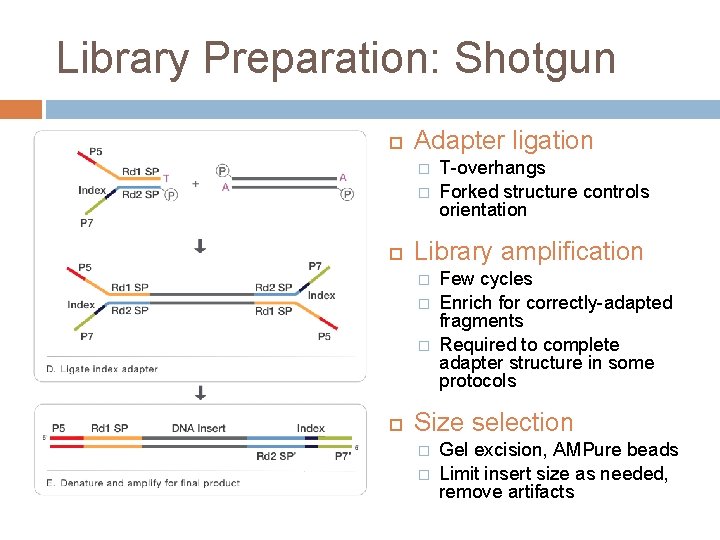
Library Preparation: Shotgun Adapter ligation � � Library amplification � � � T-overhangs Forked structure controls orientation Few cycles Enrich for correctly-adapted fragments Required to complete adapter structure in some protocols Size selection � � Gel excision, AMPure beads Limit insert size as needed, remove artifacts
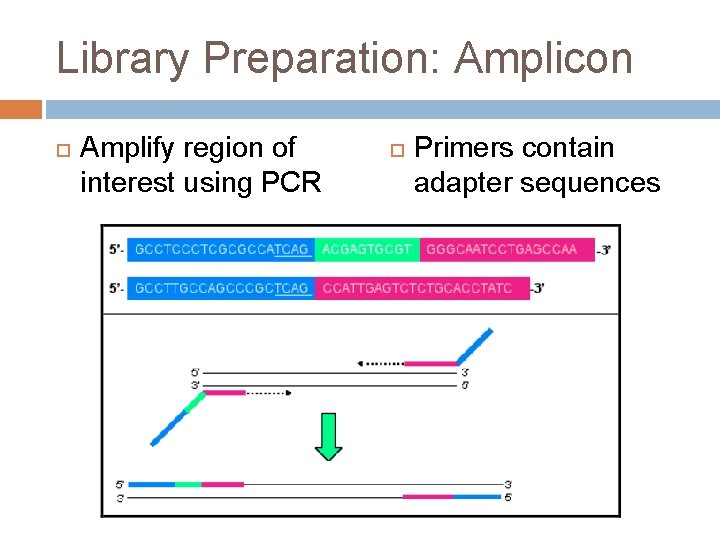
Library Preparation: Amplicon Amplify region of interest using PCR Primers contain adapter sequences
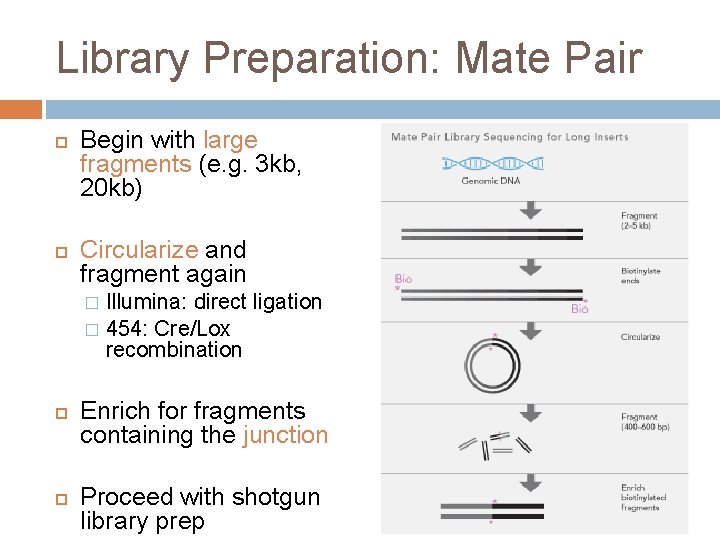
Library Preparation: Mate Pair Begin with large fragments (e. g. 3 kb, 20 kb) Circularize and fragment again Illumina: direct ligation � 454: Cre/Lox recombination � Enrich for fragments containing the junction Proceed with shotgun library prep

Library Preparation: Mate Pair Why? Paired sequences are a known distance apart; improves genome assembly Note: 454 calls these “paired end libraries”, not to be confused with Illumina’s “paired end sequencing”!

Sequencing: Illumina Cluster generation � Library fragments hybridize to oligos on the flow cell � New strand synthesized, original denatured, removed � Free end binds to adjacent oligos (bridge formation) � Complimentary strand synthesized, denatured (both tethered to flow cell) � Repeat to form clonal cluster � Cleave one oligo, denature to leave ss. DNA clusters ~800 K clusters/mm^2

Sequencing: Illumina Variety of workflows: � Single- or paired end reads � 0, 1, or 2 index reads
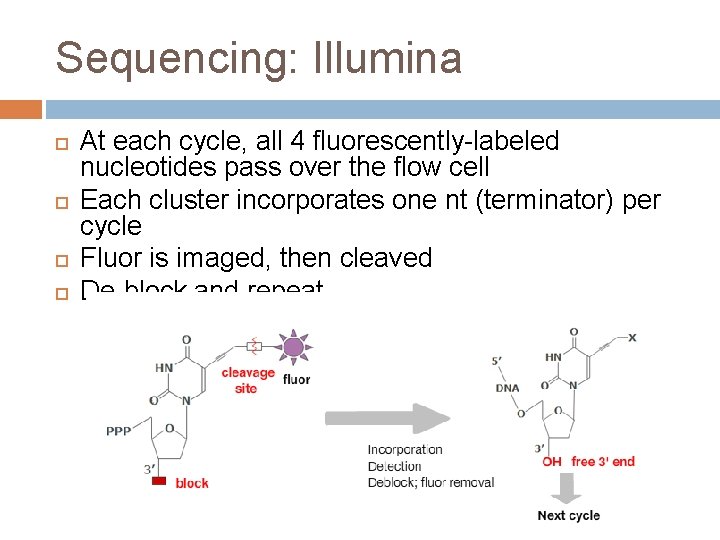
Sequencing: Illumina At each cycle, all 4 fluorescently-labeled nucleotides pass over the flow cell Each cluster incorporates one nt (terminator) per cycle Fluor is imaged, then cleaved De-block and repeat
Sequencing: Illumina Other terminology: c. Bot – accessory instrument that performs cluster generation � Lanes – divisions (8) of Hi. Seq and GAIIx flow cells � Phi. X – bacteriophage with small, balanced genome; Phi. X library spiked in with samples for QC � Phasing/pre-phasing – nt incorporation falls behind or jumps ahead on a portion of strands in the cluster and contributes to noise � Chastity filter – measures signal purity (after intensity corrections); if the background signal is high, cluster will be discarded � Base. Space – cloud computing site for processing Mi. Seq data � File format: fastq
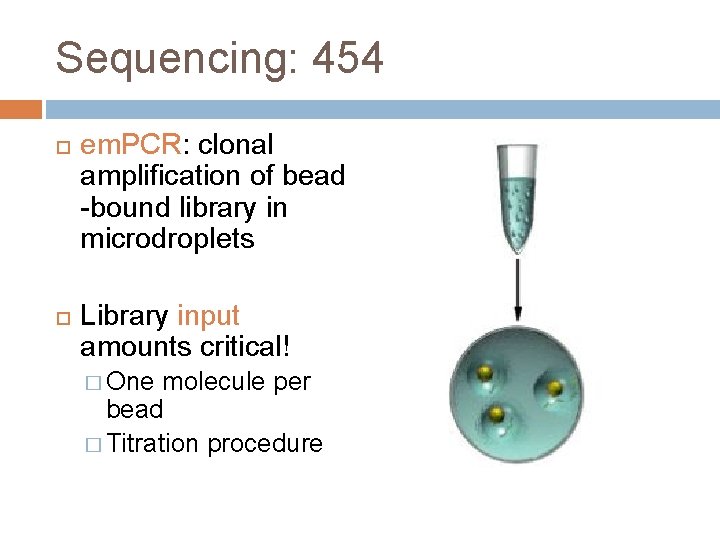
Sequencing: 454 em. PCR: clonal amplification of bead -bound library in microdroplets Library input amounts critical! � One molecule per bead � Titration procedure
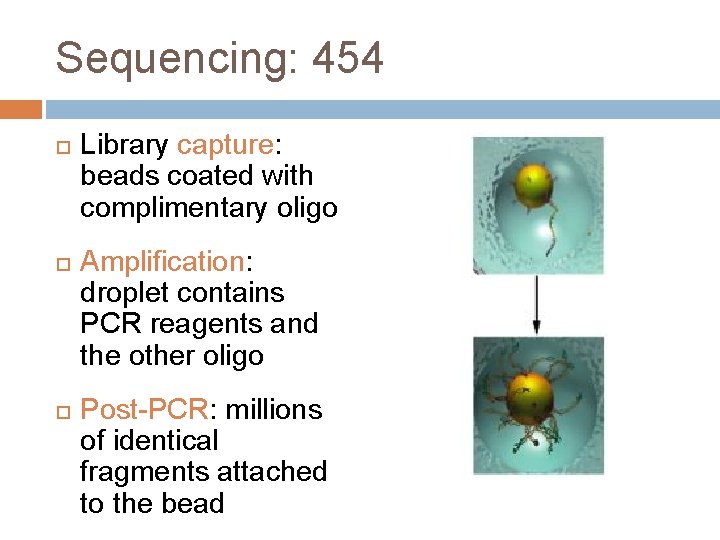
Sequencing: 454 Library capture: beads coated with complimentary oligo Amplification: droplet contains PCR reagents and the other oligo Post-PCR: millions of identical fragments attached to the bead
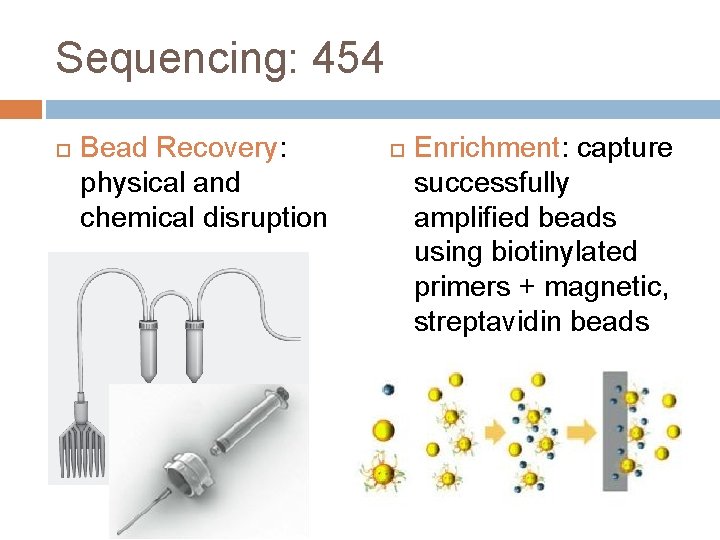
Sequencing: 454 Bead Recovery: physical and chemical disruption Enrichment: capture successfully amplified beads using biotinylated primers + magnetic, streptavidin beads
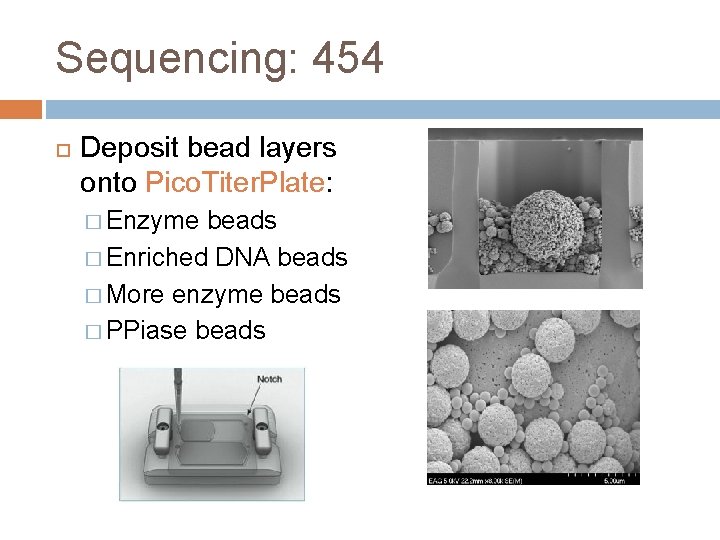
Sequencing: 454 Deposit bead layers onto Pico. Titer. Plate: � Enzyme beads � Enriched DNA beads � More enzyme beads � PPiase beads
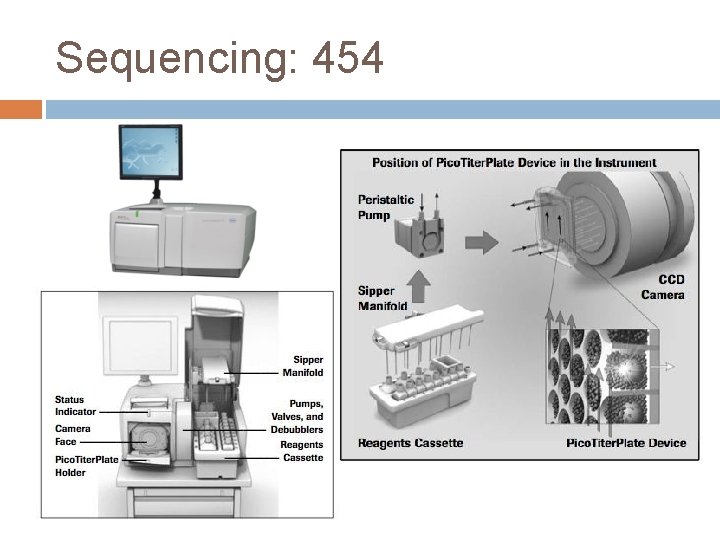
Sequencing: 454
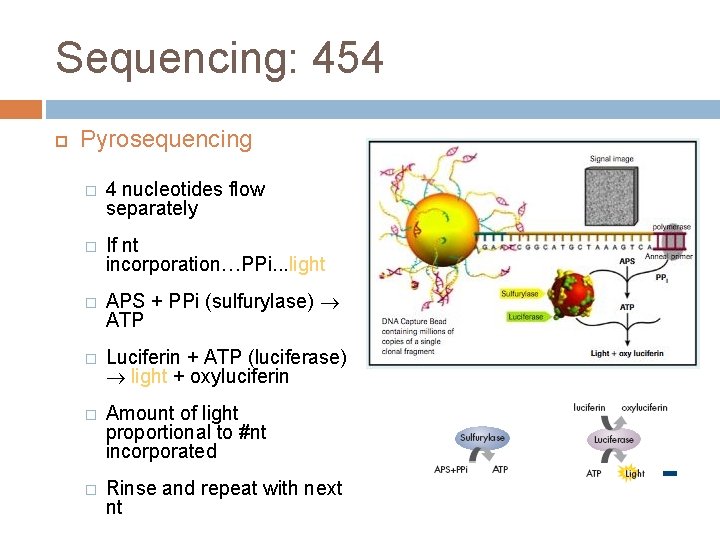
Sequencing: 454 Pyrosequencing � 4 nucleotides flow separately � If nt incorporation…PPi. . . light � APS + PPi (sulfurylase) ATP � Luciferin + ATP (luciferase) light + oxyluciferin � Amount of light proportional to #nt incorporated � Rinse and repeat with next nt
Sequencing: 454 Camera captures light emitted from every well during every nucleotide flow
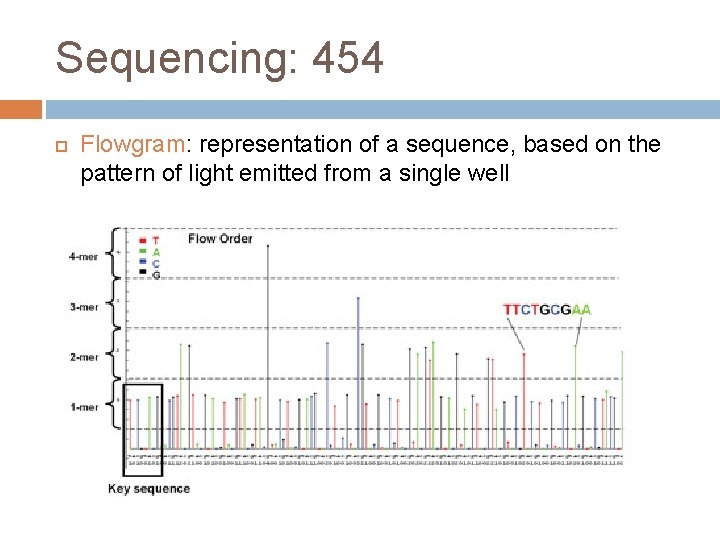
Sequencing: 454 Flowgram: representation of a sequence, based on the pattern of light emitted from a single well
Sequencing: 454 Other terminology: � Lib-L/Lib-A: adapter variants, “ligated” or “annealed” � Titanium chemistry: ~450 bp reads on all instruments � XL+ chemistry: ~700 bp reads on the FLX+ instrument � Flow: one of the four nucleotides flows over the PTP � Cycle: a set of four flows, in order � Valley flow: if number of bases incorporated in a given read during that flow is uncertain, e. g. 1. 5 units of light (background signal, homopolymers) File format: sff (standard flowgram format)
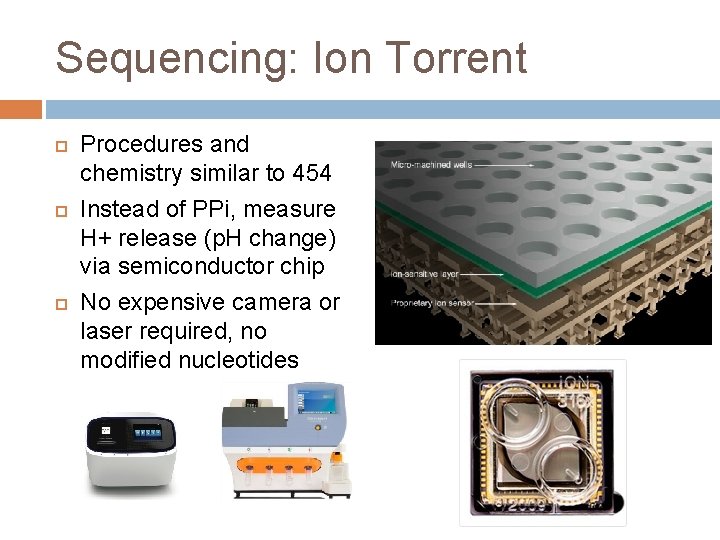
Sequencing: Ion Torrent Procedures and chemistry similar to 454 Instead of PPi, measure H+ release (p. H change) via semiconductor chip No expensive camera or laser required, no modified nucleotides

Sequence Quality Phred (Q) Score Probabilit y of Error (P) Base Call Accuracy 10 1 in 10 90% 20 1 in 100 99% 30 1 in 1 K 99. 9% 40 1 in 10 K 99. 99% 50 1 in 100 K 99. 999% Error probabilities determined using training sets, platformspecific biases Expressed as a quality value (QV or Q score) per base Similar to PHRED scores: �Q = -10 log 10 P � P = 10 -Q/10
Project 1: Microbial Genome Considerations: � Reference genome? � How much coverage do I want? � How big is the genome � How much data do I need? bp needed = genome size X coverage � Which instrument/chemistry configuration to use? Coverage � Depth (number of times a particular base is “covered” by a read (e. g. 25 X) � Breadth (% of genome with at least 1 X coverage)
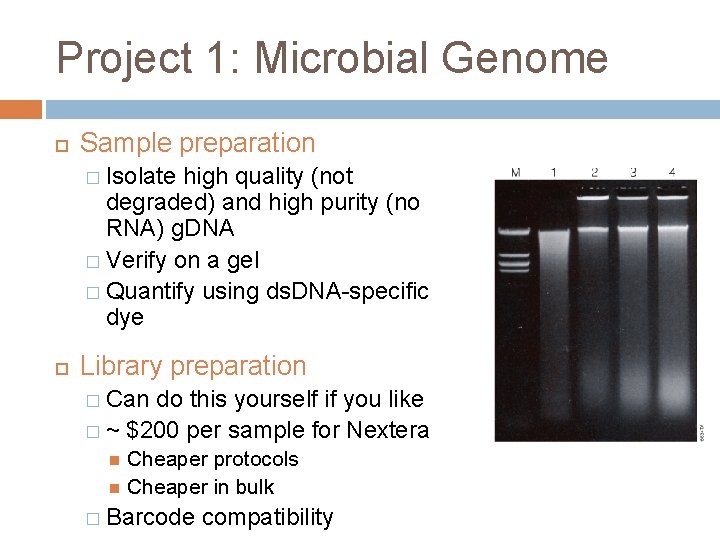
Project 1: Microbial Genome Sample preparation � Isolate high quality (not degraded) and high purity (no RNA) g. DNA � Verify on a gel � Quantify using ds. DNA-specific dye Library preparation � Can do this yourself if you like � ~ $200 per sample for Nextera Cheaper protocols Cheaper in bulk � Barcode compatibility

Project 1: Microbial Genome Library QC � Insert size confirmed on Bio. Analyzer (within range, no artifacts) � Pool barcoded libraries (normalize based on Pico. Green quantification) � Absolute quantification of library pools using q. PCR

Project 1: Microbial Genome Mi. Seq sequencing � Dilute and denature library pool (optimal concentration requires titration. . . ) � Spike in Phi. X library as needed (e. g. 1%) � Prepare and load reagents, flow cell � Basic filtering and de-multiplexing performed automatically � Download fastq files from Base. Space
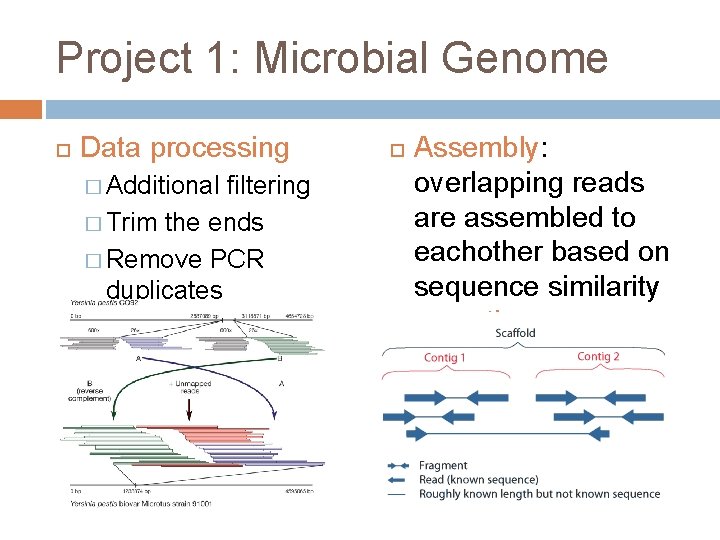
Project 1: Microbial Genome Data processing � Additional filtering � Trim the ends � Remove PCR duplicates Assembly: overlapping reads are assembled to eachother based on sequence similarity = contigs
Project 1: Microbial Genome What’s next? � Polish the genome (hybrid assemblies, mate pair libraries) � Annotate (ORFs, RNA-seq) � Compare

Project 2: Microbial Community Shotgun metagenomics � Unbiased survey of community content � Random library fragments may provide very little taxonomic resolution (e. g. conserved, unknown) � Identify genes, classify by function Targeted metagenomics � Limited survey of community content � Targeted loci provide excellent taxonomic resolution, but may exclude certain taxa � Identify OTUs, classify by taxonomy
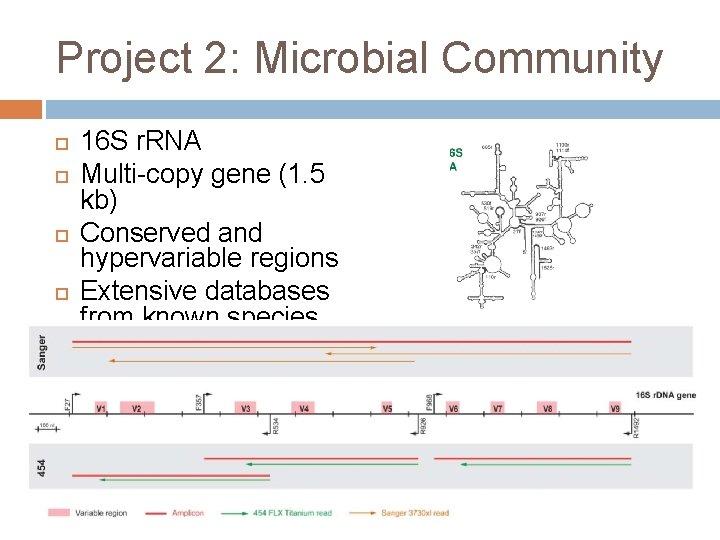
Project 2: Microbial Community 16 S r. RNA Multi-copy gene (1. 5 kb) Conserved and hypervariable regions Extensive databases from known species
Project 2: Microbial Community Considerations: � Biases in sampling methods, culturing, DNA isolation, PCR. . . replicate � Available SOPs � How many reads per sample? � Read length matters! Sample preparation: � Isolate DNA � PCR amplify, purify High-fidelity polymerase Barcoded primers No primer dimers! � Normalize PCR products and pool
Project 2: Microbial Community 454 Sequencing � em. PCR titrations with different library input � Bulk em. PCR � Sequence � Basic filtering � Collect sff files Data processing � De-multiplexing � Additional filtering � Trim the barcodes, primers � Check for chimeras
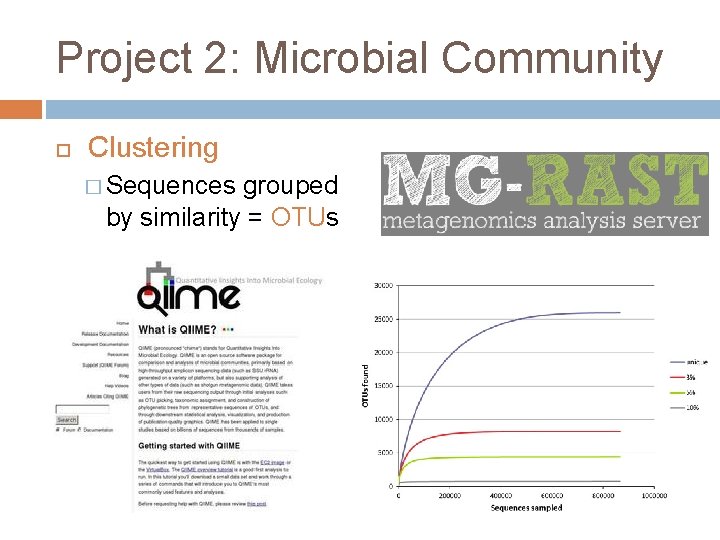
Project 2: Microbial Community Clustering � Sequences grouped by similarity = OTUs
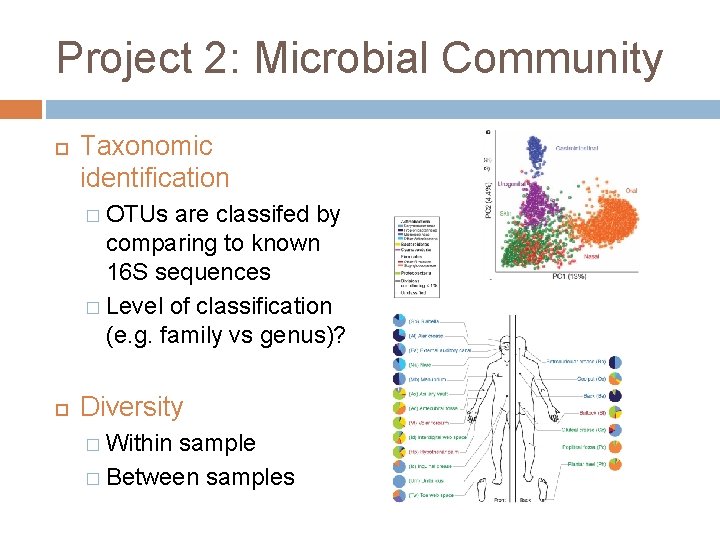
Project 2: Microbial Community Taxonomic identification � OTUs are classifed by comparing to known 16 S sequences � Level of classification (e. g. family vs genus)? Diversity � Within sample � Between samples
- Slides: 41