Nextgeneration sequencing and PBRC Next Generation Sequencer Applications
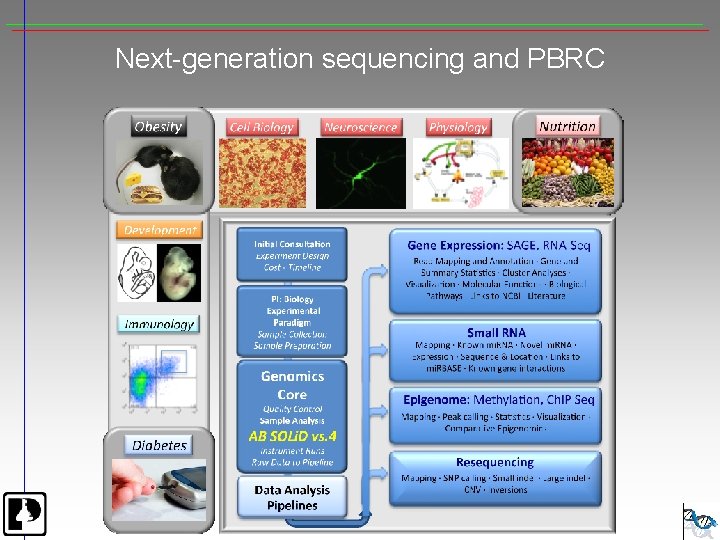
Next-generation sequencing and PBRC

Next Generation Sequencer Applications • • • De. Novo Sequencing Resequencing, Comparative Genomics Global SNP Analysis Gene Expression Analysis Methylation Studies Ch. IP Sequencing-transcription factors, histones, polymerases Transcriptome Analysis-splicing, UTRs, c. SNPs, nested transcripts Micro. RNA Discovery and quantitation Metagenomics, Microbial diversity Copy number variation Chromosomal aberrations Gene regulation studies

AB SOLi. D Ligation sequencing

How many sequence tags* do I need for my gene expression application? • • • SAGE/CAGE – 2 -5 million mappable mi. RNA – 10 million mappable Ch. IP Seq— 10 -20 million mappable Whole Transcriptome from poly. A RNA – 40 -50 million mappable Whole Transcriptome from r. RNA depleted - >50 million mappable Whole Transcriptome for Allele Specific Expression - >>50 million mappable SOLi. D™ 4 generates >1. 4 billion mappable sequences/run (2 slides) Libraries can be multiplexed to decrease the cost/sample according to the application and number of sequences needed. *For human/mouse sized genomes; smaller organisms require fewer sequence tags.

SAGE Sequencing vs. Microarray SOLi. D v 4 Microarray-Illumina Ref 8 Microarray-Illumina Ref 6 3. 6 million 25, 600 45, 200 Known and novel transcripts Known transcripts Sensitivity 6 logs 3 logs Technical Reproducibility >. 99 -. 999 0. 9 Correlation to Taqman 0. 9 0. 7 -0. 8 Multiplexing/Barcoding Yes –up to 48 RNA or 96 DNA samples No No Data Points No background –better for Hybridization process low abundance transcript creates background signal detection RNA quantity 5 -10 ug $7200 -full service 16 Sample Experiment $6100 -PI creates library Cost Hybridization process creates background signal 750 ng $3600 $5200

Primary Data Analysis - Images to bases Instrument-specific Sequences + Quality values Secondary Data Analysis – Bases to alignments/contigs Applications • Tag Profiling • Small RNA Analysis • Transcriptome seq. • Ch. IP-Seq • Methylation Analysis • Resequencing • De novo assembly One or more Data sets Discovery Ref Seq + Alignment Assembly, De Novo Sample/Library Quality Run Quality Bioinformatics: Geospiza Algorithms • Eland • Maq • SOAP • Velvet • Newbler • Mapreads • Others … Tertiary Data Analysis – Experiment Specific • Differential expression • Methylation sites • Binding sites • Gene association • Genomic structure

Next-gen sequencing: applications – Genome analysis: basic and translational research • Genetics of disease – new frontiers • Exome resequencing: confirmation of GWAS • Genome sequence as diagnostic tool • Genetic counseling – Epigenome analysis: basic research; biomarkers • Analyses of DNA methylation, transcription factors, histone modifications, non-coding RNA • Epigenomic biomarkers of disease – Gene expression analysis: basic research; diagnostics & biomarkers • Whole transcriptome: all transcribed sequences in a cell • SAGE analysis: expression of known genes • Small RNA: micro. RNA as regulators of biology – Genotype to phenotype: a new frontier • Pathology: systems biology • Diagnosis: data filtering • Personalized Genomic Medicine: Treatment recommendations

Next-gen sequencing: challenges – Rapid growth in methodology • Technology and equipment changes & upgrades – High demands on informatics: • Staff • Software • Computational resources – New ways of handling data needed: • Interpretation • Publication • Storage
- Slides: 8