New Primer Sets for Y Chromosome and CODIS
- Slides: 1

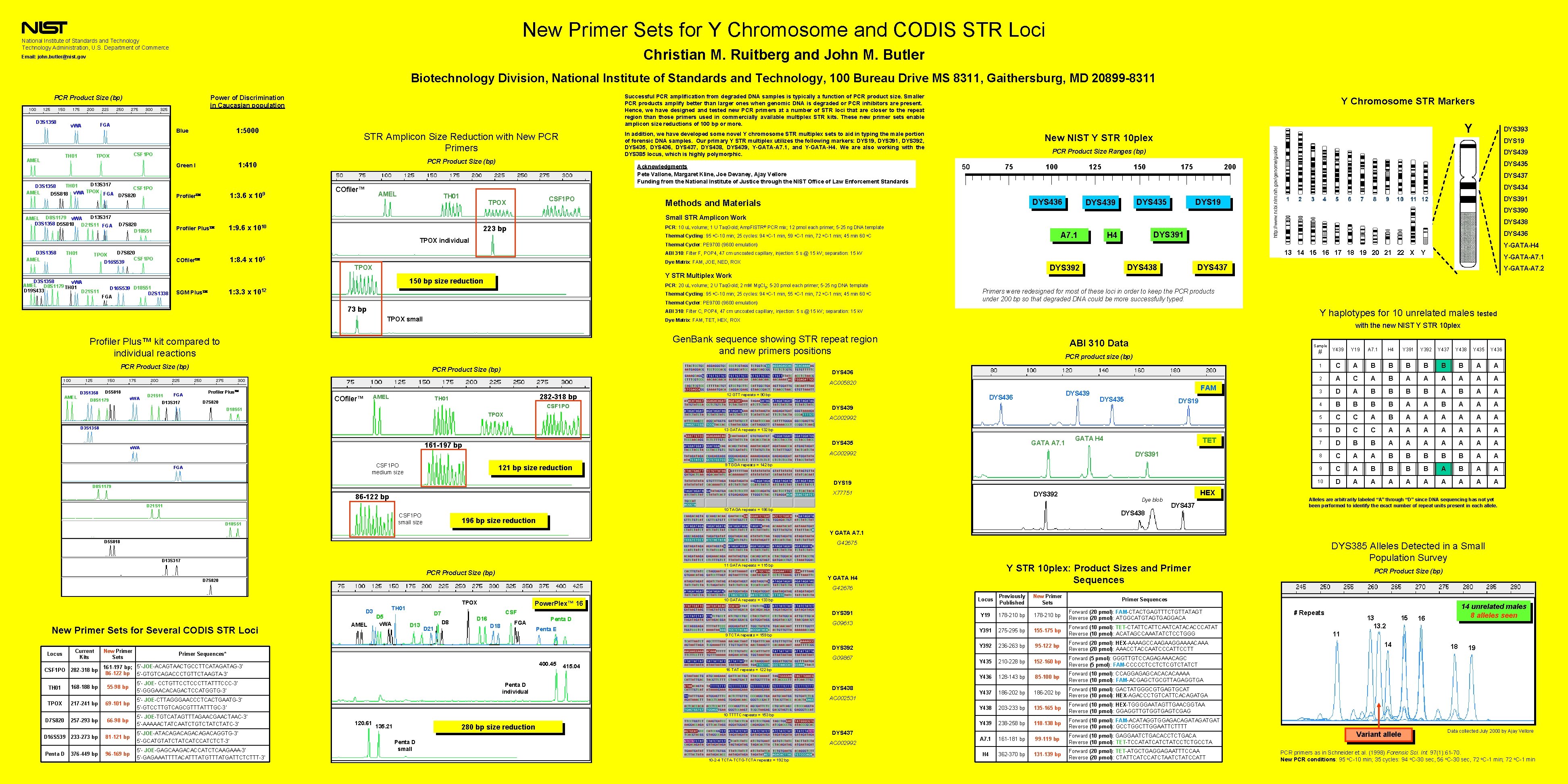
New Primer Sets for Y Chromosome and CODIS STR Loci National Institute of Standards and Technology Administration, U. S. Department of Commerce Christian M. Ruitberg and John M. Butler Email: john. butler@nist. gov Biotechnology Division, National Institute of Standards and Technology, 100 Bureau Drive MS 8311, Gaithersburg, MD 20899 -8311 FGA v. WA TH 01 AMEL 1: 5000 Blue CSF 1 PO TPOX D 13 S 317 TH 01 D 3 S 1358 CSF 1 PO TPOX v. WA AMEL D 5 S 818 FGA D 7 S 820 D 13 S 317 AMEL D 8 S 1179 v. WA D 3 S 1358 D 5 S 818 D 21 S 11 FGA 1: 3. 6 x 109 Profiler In addition, we have developed some novel Y chromosome STR multiplex sets to aid in typing the male portion of forensic DNA samples. Our primary Y STR multiplex utilizes the following markers: DYS 19, DYS 391, DYS 392, DYS 435, DYS 436, DYS 437, DYS 438, DYS 439, Y-GATA-A 7. 1, and Y-GATA-H 4. We are also working with the DYS 385 locus, which is highly polymorphic. STR Amplicon Size Reduction with New PCR Primers PCR Product Size (bp) 1: 410 Green I COfiler™ AMEL TH 01 Acknowledgments Pete Vallone, Margaret Kline, Joe Devaney, Ajay Vellore Funding from the National Institute of Justice through the NIST Office of Law Enforcement Standards CSF 1 PO TPOX D 7 S 820 D 18 S 51 1: 9. 6 x 1010 Profiler Plus D 7 S 820 TPOX CSF 1 PO D 16 S 539 TH 01 D 3 S 1358 v. WA AMEL D 8 S 1179 TH 01 D 19 S 433 D 21 S 11 New NIST Y STR 10 plex PCR Product Size Ranges (bp) 50 75 100 125 DYS 436 Methods and Materials 150 175 DYS 439 200 DYS 19 PCR: 10 u. L volume; 1 U Taq. Gold; Amp. Fl. STR® PCR mix; 12 pmol each primer; 5 -25 ng DNA template 223 bp PCR: 20 u. L volume; 2 U Taq. Gold; 2 m. M Mg. Cl 2; 5 -20 pmol each primer; 5 -25 ng DNA template Thermal Cycling: 95 o. C-10 min; 25 cycles: 94 o. C-1 min, 55 o. C-1 min, 72 o. C-1 min; 45 min 60 o. C Thermal Cycler: PE 9700 (9600 emulation) 73 bp DYS 438 DYS 392 Y STR Multiplex Work 1: 3. 3 x 1012 TPOX small AMEL D 8 S 1179 v. WA D 21 S 11 FGA PCR Product Size (bp) D 13 S 317 AMEL COfiler™ D 7 S 820 282 -318 bp ABI 310 Data DYS 439 DYS 436 FAM DYS 435 DYS 19 DYS 439 TPOX AC 002992 D 3 S 1358 13 GATA repeats = 132 bp GATA A 7. 1 DYS 435 161 -197 bp AC 002992 CSF 1 PO medium size 121 bp size reduction GATA H 4 TET DYS 391 9 TGGA repeats = 142 bp DYS 19 D 8 S 1179 X 77751 86 -122 bp D 21 S 11 DYS 392 10 TAGA repeats = 186 bp CSF 1 PO small size D 18 S 51 3 4 5 6 7 8 9 DYS 391 10 11 12 DYS 390 DYS 438 DYS 436 Y Y-GATA-A 7. 1 Y-GATA-A 7. 2 Sample PCR product size (bp) 12 GTT repeats = 90 bp CSF 1 PO D 18 S 51 FGA 2 # Y 439 Y 19 A 7. 1 H 4 Y 391 Y 392 Y 437 Y 438 Y 435 Y 436 1 C A B B B A A 2 A C A B A A A 3 D A B B A B A A 4 B B A A A 5 C C A B A A A 6 D C C A A A A 7 D B B A A A A 8 C A A B B B A A 9 C A B B A B A A 10 D A A A A A DYS 436 TH 01 v. WA 1 with the new NIST Y STR 10 plex Gen. Bank sequence showing STR repeat region and new primers positions Profiler Plus DYS 434 Y haplotypes for 10 unrelated males tested AC 005820 D 5 S 818 DYS 437 Primers were redesigned for most of these loci in order to keep the PCR products under 200 bp so that degraded DNA could be more successfully typed. Dye Matrix: FAM, TET, HEX, ROX Profiler Plus™ kit compared to individual reactions D 3 S 1358 DYS 435 DYS 437 ABI 310: Filter C, POP 4, 47 cm uncoated capillary, injection: 5 s @ 15 k. V; separation: 15 k. V PCR Product Size (bp) DYS 439 13 14 15 16 17 18 19 20 21 22 X Dye Matrix: FAM, JOE, NED, ROX 150 bp size reduction SGM Plus DYS 19 Y-GATA-H 4 ABI 310: Filter F, POP 4, 47 cm uncoated capillary, injection: 5 s @ 15 k. V; separation: 15 k. V TPOX D 16 S 539 D 18 S 51 D 2 S 1338 FGA DYS 391 H 4 DYS 393 Thermal Cycler: PE 9700 (9600 emulation) 1: 8. 4 x 105 COfiler A 7. 1 Thermal Cycling: 95 o. C-10 min; 25 cycles: 94 o. C-1 min, 59 o. C-1 min, 72 o. C-1 min; 45 min 60 o. C Y Human Genome Small STR Amplicon Work TPOX individual D 3 S 1358 AMEL Y Chromosome STR Markers http: //www. ncbi. nlm. nih. gov/genome/guide/ D 3 S 1358 Successful PCR amplification from degraded DNA samples is typically a function of PCR product size. Smaller PCR products amplify better than larger ones when genomic DNA is degraded or PCR inhibitors are present. Hence, we have designed and tested new PCR primers at a number of STR loci that are closer to the repeat region than those primers used in commercially available multiplex STR kits. These new primer sets enable amplicon size reductions of 100 bp or more. Power of Discrimination in Caucasian population PCR Product Size (bp) HEX Dye blob DYS 437 Alleles are arbitrarily labeled “A” through “D” since DNA sequencing has not yet been performed to identify the exact number of repeat units present in each allele. DYS 438 196 bp size reduction Y GATA A 7. 1 D 5 S 818 G 42675 D 13 S 317 Y STR 10 plex: Product Sizes and Primer Sequences 11 GATA repeats = 115 bp PCR Product Size (bp) Y GATA H 4 D 7 S 820 G 42676 D 3 New Primer Sets for Several CODIS STR Loci Locus CSF 1 PO TH 01 Current Kits 282 -318 bp 168 -188 bp New Primer Sets TH 01 D 5 v. WA CSF D 7 D 13 D 21 Power. Plex™ 16 D 8 D 16 D 18 FGA DYS 391 Penta D Penta E 9 TCTA repeats = 159 bp Primer Sequences* 400. 45 161 -197 bp; 5’-JOE-ACAGTAACTGCCTTCATAG-3’ 86 -122 bp 5’-GTGTCAGACCCTGTTCTAAGTA-3’ 55 -98 bp 5’- JOE- CCTGTTCCTCCCTTATTTCCC-3’ 5’-GGGAACACAGACTCCATGGTG-3’ TPOX 217 -241 bp 69 -101 bp D 7 S 820 257 -293 bp 66 -98 bp 5’- JOE-TGTCATAGTTTAGAACTAAC-3’ 5’-AAAAACTATCAATCTGTCTATC-3’ D 16 S 539 233 -273 bp 81 -121 bp 5’-JOE-ATACAGACAGGTG-3’ 5’-GCATGTATCATCCATCTCT-3’ Penta D 96 -169 bp 5’- JOE-GAGCAAGACACCATCTCAAGAAA-3’ 5’-GAGAAATTTTACATTTATGATTCTCTTT-3’ G 09867 415. 04 Previously Published New Primer Sets Y 19 178 -210 bp Y 391 275 -295 bp 155 -175 bp Forward (10 pmol): TET-CTATTCAATCATACACCCATAT Reverse (10 pmol): ACATAGCCAAATATCTCCTGGG Y 392 236 -263 bp 95 -122 bp Forward (20 pmol): HEX-AAAAGCCAAGAAGGAAAACAAA Reverse (20 pmol): AAACCTACCAATCCCATTCCTT Y 435 210 -228 bp 152 -160 bp Forward (5 pmol): GGGTTGTCCAGAGAAACAGC Reverse (5 pmol): FAM-CCCCCTCCTCTCGTCTATCT Y 436 128 -143 bp 85 -100 bp Forward (10 pmol): CCAGGAGAGCACAAAA Reverse (10 pmol): FAM-ACGAGCTGCGTTAGAGGTGA Y 437 186 -202 bp Forward (10 pmol): GACTATGGGCGTGAGTGCAT Reverse (10 pmol): HEX-AGACCCTGTCATTCACAGATGA Y 438 203 -233 bp 135 -165 bp Forward (10 pmol): HEX-TGGGGAATAGTTGAACGGTAA Reverse (10 pmol): GGAGGTTGTGGTGAGTCGAG Y 439 238 -258 bp 118 -138 bp Forward (10 pmol): FAM-ACATAGGTGGAGACAGATGAT Reverse (10 pmol): GCCTGGCTTGGAATTCTTTT 16 TAT repeats = 122 bp Penta D individual DYS 438 AC 002531 10 TTTTC repeats = 153 bp 120. 61 280 bp size reduction 135. 21 DYS 437 Penta D small AC 002992 10 -2 -4 TCTA-TCTG-TCTA repeats = 192 bp PCR Product Size (bp) Primer Sequences Forward (20 pmol): FAM-CTACTGAGTTTCTGTTATAGT Reverse (20 pmol): ATGGCATGTAGTGAGGACA G 09613 DYS 392 5’- JOE-CTTAGGGAACCCTCACTGAATG-3’ 5’-GTCCTTGTCAGCGTTTATTTGC-3’ 376 -449 bp AMEL TPOX Locus 10 GATA repeats = 133 bp DYS 385 Alleles Detected in a Small Population Survey A 7. 1 161 -181 bp 99 -119 bp Forward (10 pmol): GAGGAATCTGACACCTCTGACA Reverse (10 pmol): TET-TCCATATCATCTATCCTCTGCCTA H 4 362 -370 bp 131 -139 bp Forward (20 pmol): TET-ATGCTGAGGAGAATTTCCAA Reverse (20 pmol): CTATTCATCTAATCTATCCATT # Repeats 13 13. 2 15 14 unrelated males 8 alleles seen 16 11 14 Variant allele 18 19 Data collected July 2000 by Ajay Vellore PCR primers as in Schneider et al. (1998) Forensic Sci. Int. 97(1): 61 -70. New PCR conditions: 95 o. C-10 min; 35 cycles: 94 o. C-30 sec, 56 o. C-30 sec, 72 o. C-1 min; 72 o. C-1 min