NEW APPROACHES TO PROTEIN STRUCTURE PREDICTION AND DESIGN






- Slides: 43

NEW APPROACHES TO PROTEIN STRUCTURE PREDICTION AND DESIGN Joe De. Bartolo

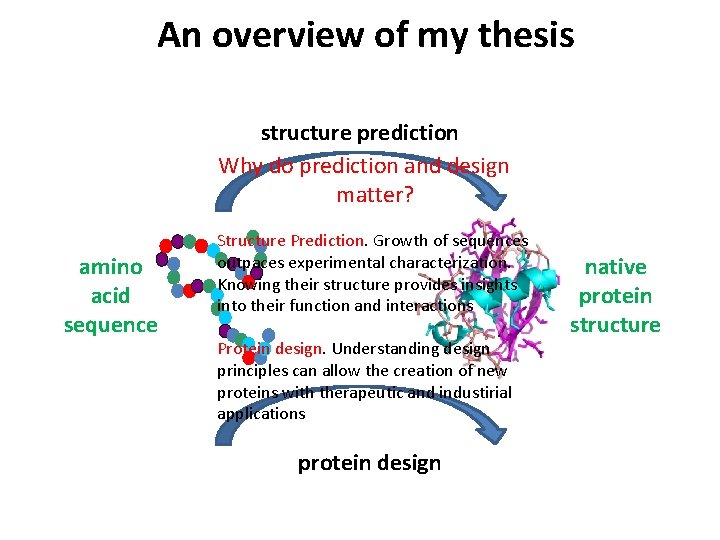
An overview of my thesis structure prediction Why do prediction and design matter? amino acid sequence Structure Prediction. Growth of sequences outpaces experimental characterization. Knowing their structure provides insights into their function and interactions Protein design. Understanding design principles can allow the creation of new proteins with therapeutic and industirial applications protein design native protein structure

Protein structure prediction and design PART III PART IV It. Fix: Homology-free structure prediction SPEED: It. Fix enhanced with evolution Future directions in prediction Protein design

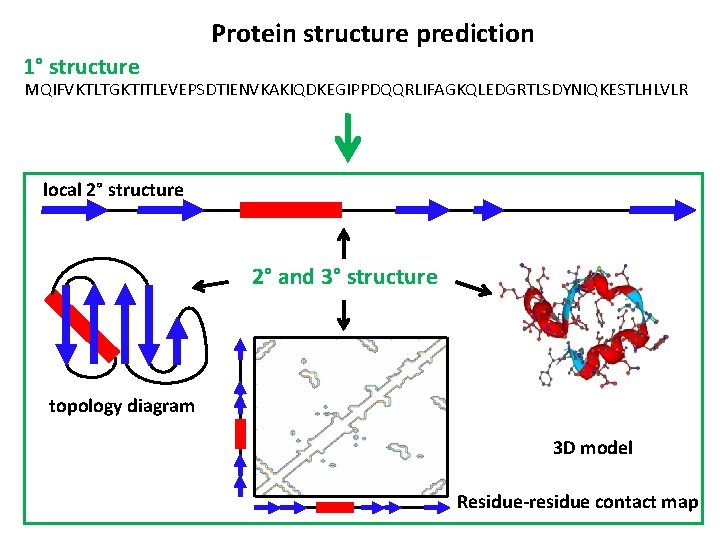
Protein structure prediction 1° structure MQIFVKTLTGKTITLEVEPSDTIENVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLSDYNIQKESTLHLVLR local 2° structure 2° and 3° structure topology diagram 3 D model Residue-residue contact map

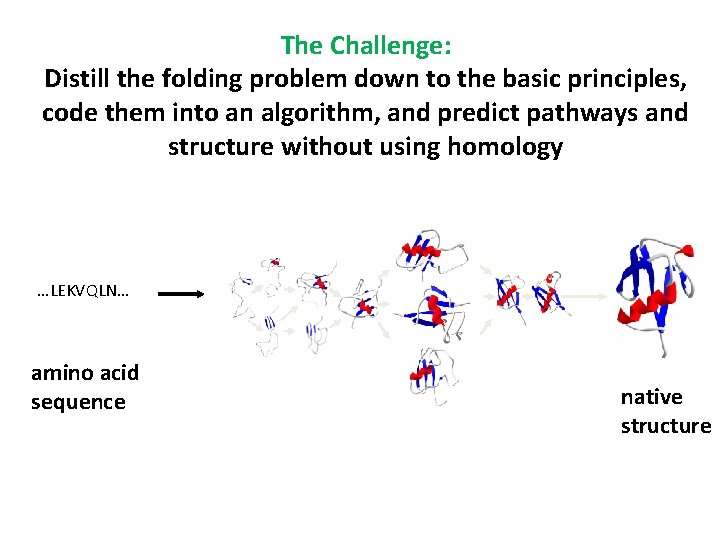
The Challenge: Distill the folding problem down to the basic principles, code them into an algorithm, and predict pathways and structure without using homology …LEKVQLN… amino acid sequence native structure

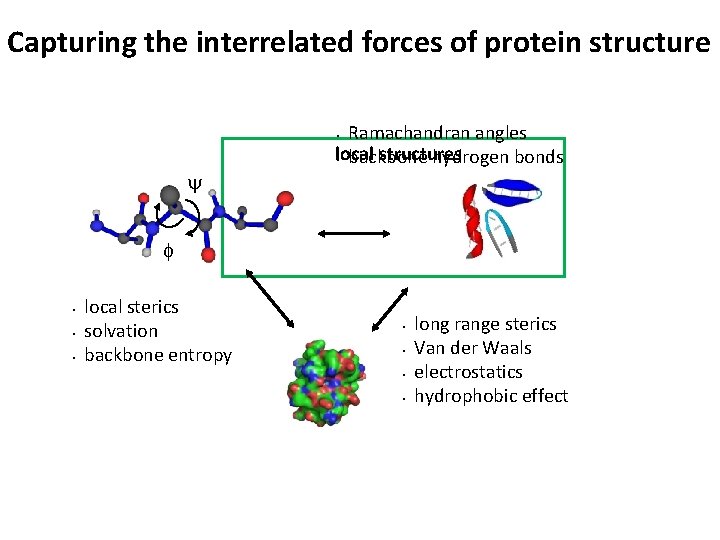
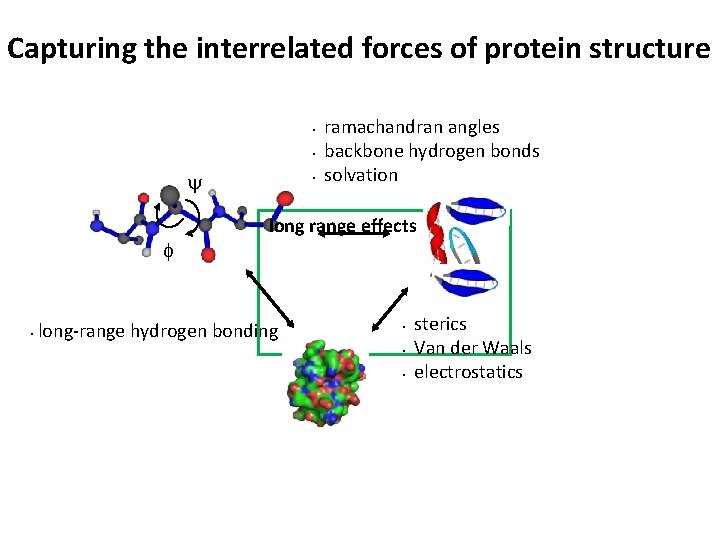
Capturing the interrelated forces of protein structure Ramachandran angles local structures • backbone hydrogen bonds • y f • • • local sterics solvation backbone entropy • • long range sterics Van der Waals electrostatics hydrophobic effect

The overlapping features of local protein structure turn β-strand α-helix backbone 180 Ramachandran torsion ψ angles y f -180 180 φ -180 φ 180 -180 φ backbone H-bonds polar amphipathic sidechain patterning apolar mostly polar 180

Capturing the interrelated forces of protein structure • • y f • • ramachandran angles backbone hydrogen bonds solvation long range effects long-range hydrogen bonding • • • sterics Van der Waals electrostatics

3° packing specificity of the chain hydrophobic effect surface residue placement solvent exposed residues salt bridges and other favorable pairings apolar buried residues long-range hydrogen bonding contacts that are highly separated in sequence

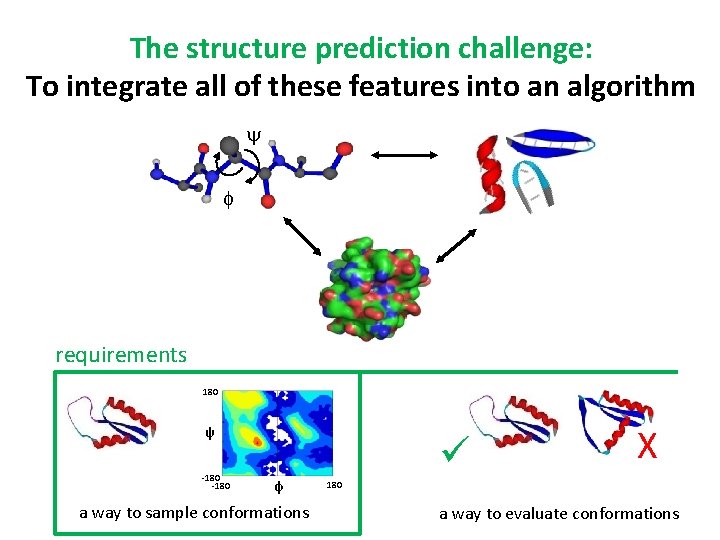
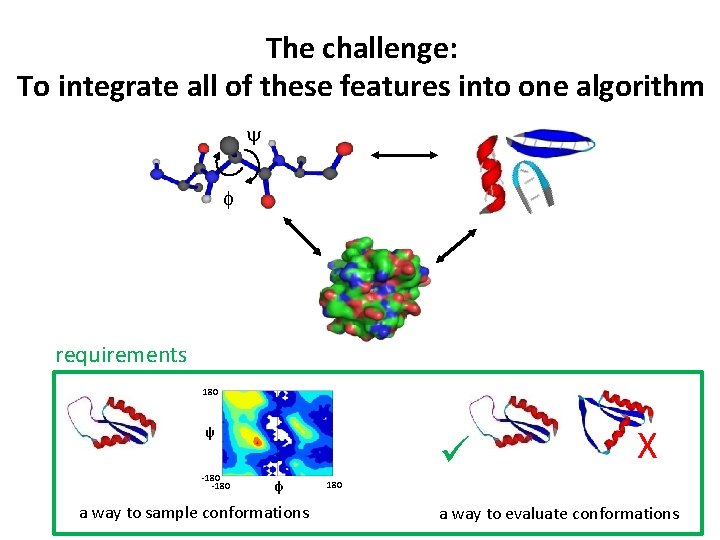
The structure prediction challenge: To integrate all of these features into an algorithm y f requirements 180 ψ -180 φ a way to sample conformations X 180 a way to evaluate conformations

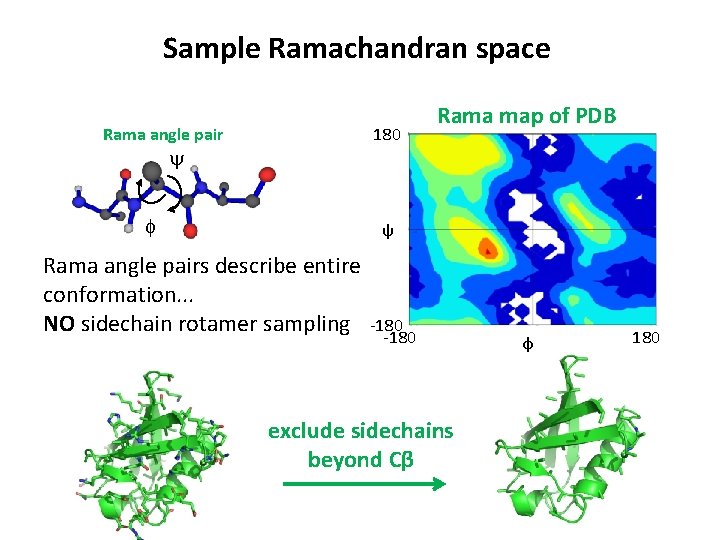
Sample Ramachandran space Rama angle pair 180 Rama map of PDB y f ψ Rama angle pairs describe entire conformation. . . NO sidechain rotamer sampling -180 exclude sidechains beyond Cβ φ 180

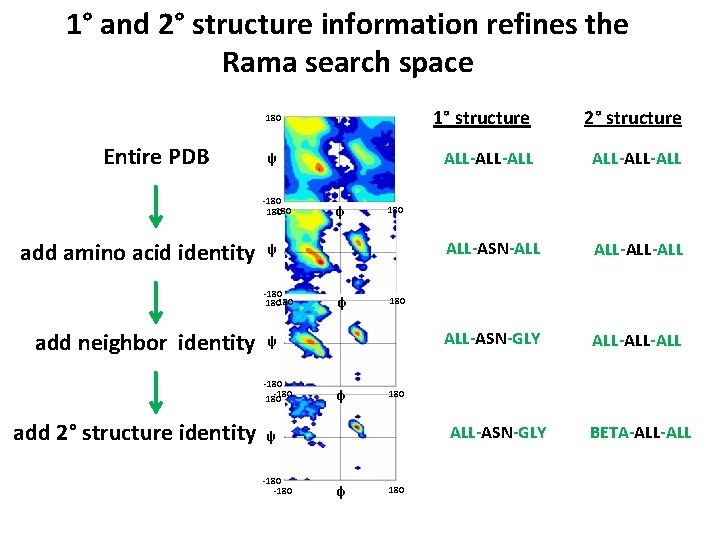
1° and 2° structure information refines the Rama search space 180 Entire PDB ψ -180 180 add amino acid identity φ ALL-ALL ALL-ASN-ALL ALL-ALL ALL-ASN-GLY ALL-ALL 180 ALL-ASN-GLY ψ -180 ALL-ALL 180 ψ -180 180 add 2° structure identity φ 2° structure 180 ψ -180 180 add neighbor identity φ 1° structure φ 180 BETA-ALL

The structure prediction challenge: To integrate all of these features into one algorithm y f requirements 180 ψ -180 φ a way to sample conformations X 180 a way to evaluate conformations

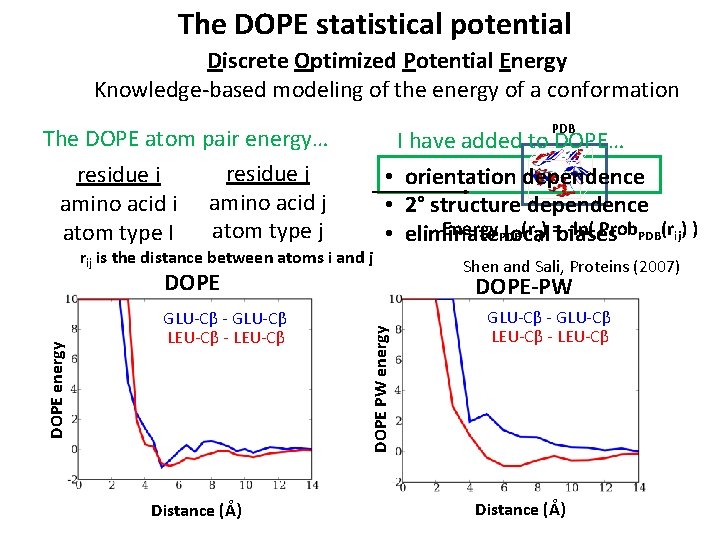
The DOPE statistical potential Discrete Optimized Potential Energy Knowledge-based modeling of the energy of a conformation The DOPE atom pair energy… residue j residue i amino acid j atom type I PDB I have added to DOPE… rij is the distance between atoms i and j • orientation dependence • 2° structure dependence Energy. PDB (rij) =biases -ln( Prob. PDB(rij) ) • eliminate local GLU-Cβ - GLU-Cβ LEU-Cβ - LEU-Cβ Distance (Å) DOPE-PW DOPE PW energy DOPE Shen and Sali, Proteins (2007) GLU-Cβ - GLU-Cβ LEU-Cβ - LEU-Cβ Distance (Å)

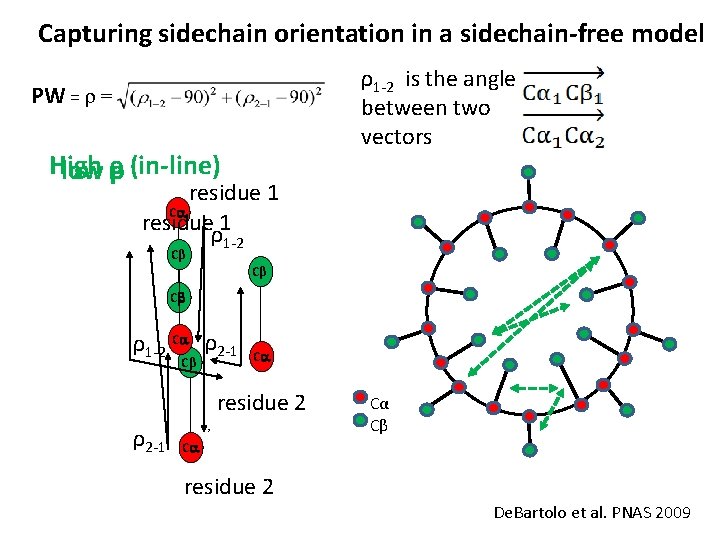
Capturing sidechain orientation in a sidechain-free model ρ1 -2 is the angle between two vectors PW = r = High low ρρ (in-line) residue 1 Ca residue 1 ρ1 -2 Cβ Cβ Cb ρ1 -2 Ca ρ2 -1 Cβ Ca residue 2 ρ2 -1 , Cα Cβ Ca residue 2 De. Bartolo et al. PNAS 2009

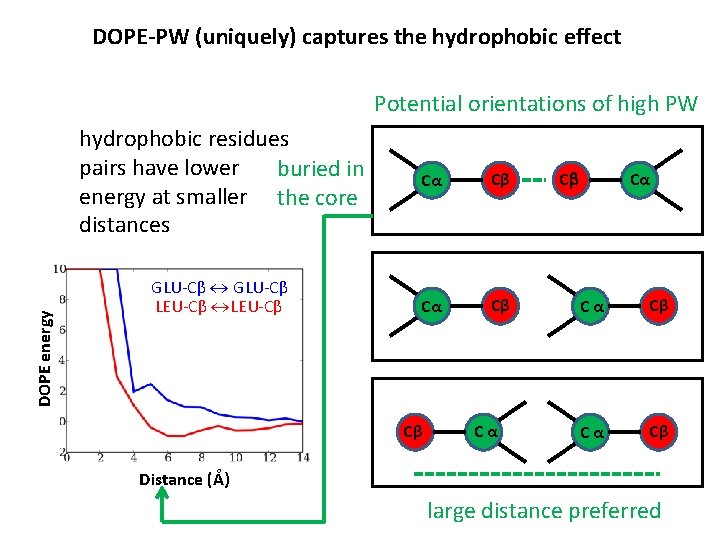
DOPE-PW (uniquely) captures the hydrophobic effect Potential orientations of high PW DOPE energy hydrophobic residues pairs have lower buried in energy at smaller the core distances GLU-Cβ LEU-Cβ Cα Cβ Cβ Cα Cb Cα Cα Cβ Distance (Å) large distance preferred

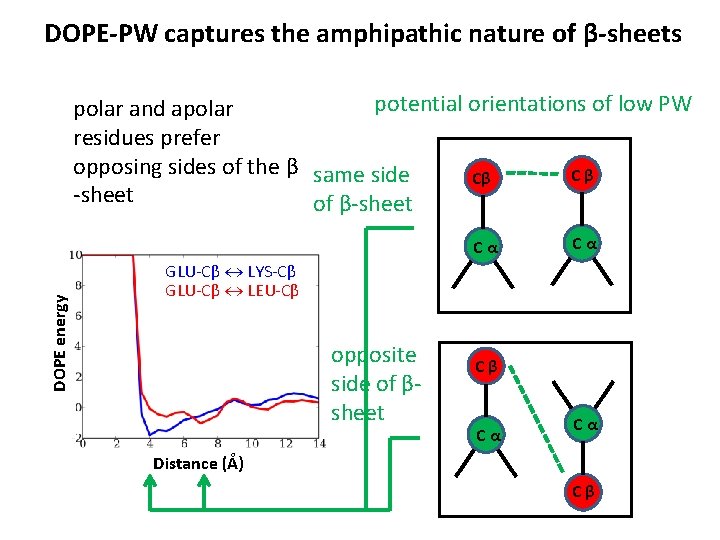
DOPE-PW captures the amphipathic nature of β-sheets potential orientations of low PW polar and apolar residues prefer opposing sides of the β same side Cβ Cβ -sheet of β-sheet DOPE energy Cα Cα GLU-Cβ LYS-Cβ GLU-Cβ LEU-Cβ opposite side of βsheet Cβ Cα Cα Distance (Å) Cβ

The challenge: To integrate all of these features into one algorithm y f requirements 180 ψ -180 φ a way to sample conformations X 180 a way to evaluate conformations

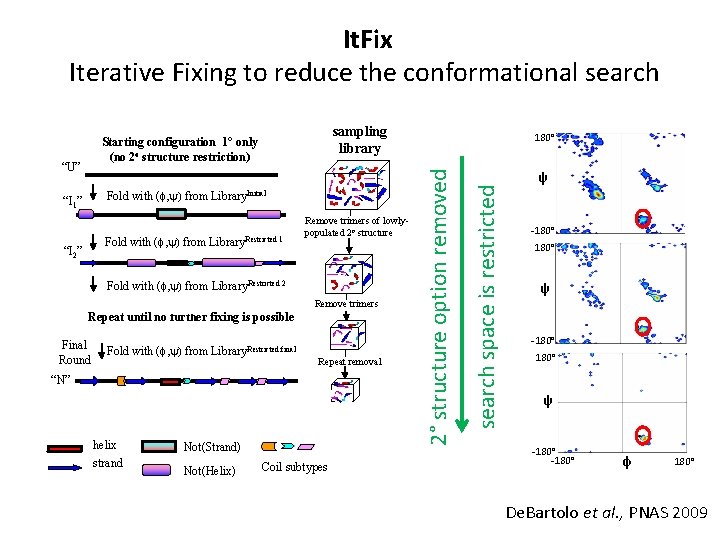
It. Fix Iterative Fixing to reduce the conformational search sampling library Fold with (f, y) from Library. Initial “I 1” Fold with (f, y) from Library. Restricted 1 “I 2” Remove trimers of lowlypopulated 2 o structure Fold with (f, y) from Library. Restricted 2 Remove trimers Repeat until no further fixing is possible Final Round Fold with (f, y) from Library. Restricted final Repeat removal “N” helix strand Not(Strand) Not(Helix) Coil subtypes search space is restricted “U” 180° 2° structure option removed Starting configuration 1° only (no 2 o structure restriction) ψ -180° ψ -180° φ 180° De. Bartolo et al. , PNAS 2009

Homology-free It. Fix 2° and 3° structure prediction results Native It. Fix SSPro PSIPRED ---HHHHHHHH-----GGGHHHHHHHHT---HHHHH-TT-THHHH---HHHHHHHHT-----S-HHHHHHHHT-S--HHHHHT---HHHHHHHHHHHHHHE-TTHHHHHHHHT--HHHHHT-TTHHHHH---HHHHHHHH-----HHHHHHHHH----HHHHHHHHH-- Native It. Fix SSPro PSIPRED -HHHHHHTT-SS--HHHHHHT--HHHHHHHH--HHHHHH-----HHHHHH--S-HHHHHHHHHHHHHHHHHHHHHHH-HHHHEEHEHHHHHHH--HHHHHHH-----HHHHHHHHHHHH-HHHH--- 1 b 72 1. 6 Å Native It. Fix SSPro Psi. Pred -EEEEETTTTEEEEE-TTS--EEEEGGGB-SSSS----TT-EEEEETTEEEEE--EEEE-STTTEEEEEEET-T-EEEEEEE--SSS-----TS--EEEEEEES--S----EEEEE--TEEEEEE-TTTTEEEE--TT--EEEEEEEHEETTT--E--TT-EEEE-TT--E-EE------EEEE----EEEEE-----EEEEEEE-------EEEE-----EEEEEE--- 1 csp 6. 0 Å Native It. Fix SSPro PSIPRED --BGGG---SEEEEE-TTS-EEEEEEHHHHHTT-EEEEEETTSSS-EEEEE-SSSSEEEEEE-TTS-EEEEEEHHHHHHT--EEEE-TTSSS-EEEEE--BBTEEE-EEEEEEETTT-EEEEE-HHHHHHT--EEEE-TT----EEEE------EEEEE-----EEEEE-HHHHH----EEEE-------EEEE-- 1 tif 4. 2 Å Native It. Fix SSPro Psi. Pred -HHHHHHTT--HHHHTS-HHHHTTS-SS-TTHHHHHHHTT--HHHHHHHHHHHHT--HHHHHHHHTT--SS----HHHHHHHT--HHHHH---HHHHHHHHHHHT-HHHHHTT-------HHHHHT--HHHHHHHHHHH----HHHHHHHH------HHHHHH---HHHH-- 1 r 69 2. 4 Å Native It. Fix SSPro PSIPRED -EEEEEETTS-EEEEE--TTSBHHHHHH---GGGEEEEETTEE--TTSBTGGGT--TT-EEEEEETTS-EEEEEE---S-B-HHHHHSS---SSEEEEETT----TT-B-----EEEEEE-EEEEEEETTEEEEEEE---SHHHHHHTTT---T--E--ETT-E--TT-EEEEEE-EEEEEE-----HHHHHH---HHHEEEEE------HHH-------EEEEEE- 1 af 7 2. 5 Å 1 ubq 3. 1 Å De. Bartolo et al. , PNAS 2009

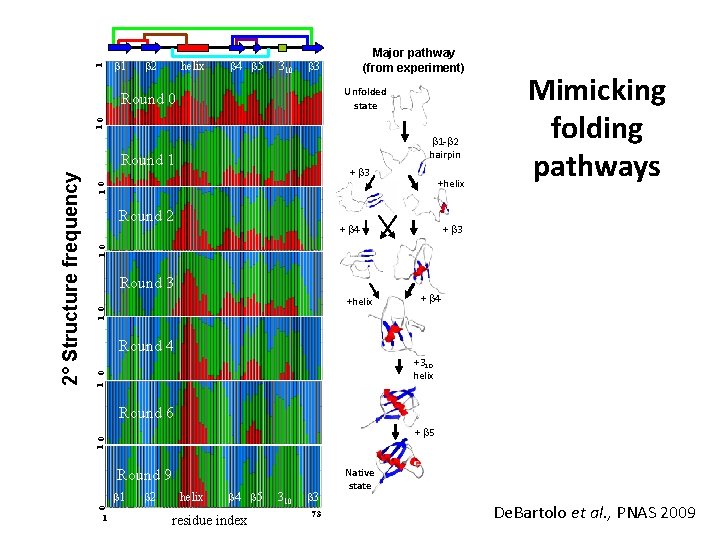
1 b 2 helix b 4 b 5 310 Major pathway (from experiment) b 3 Unfolded state 10 Round 0 b 1 -b 2 hairpin + b 3 +helix 10 Round 2 + b 4 + b 3 1 0 Round 3 1 0 +helix + b 4 Round 4 1 0 +310 helix Round 6 10 + b 5 Round 9 b 1 b 2 helix b 4 b 5 0 2° Structure frequency Round 1 Mimicking folding pathways 1 residue index 310 b 3 73 Native state De. Bartolo et al. , PNAS 2009

Part I Conclusions Challenge: Distill the folding problem down to the basic principles, code them into an algorithm, and predict pathways and structure without using homology What novel about how we approached this challenge? Use basic principles of protein structure and folding. Search strategies: mimic true folding behavior i) Coupled 2° & 3° structure formation ii) Iterative fixing to reduce the search iii) Outputs pathway information Energy functions: orientational and 2° structure dependence

Protein structure prediction and design PART III PART IV It. Fix: Homology-free structure prediction SPEED: It. Fix enhanced with evolution Future directions in prediction Protein design ψ φ Cover image of Protein Science, March 2010

SPEED: Structure Prediction Enhanced by Evolutionary Diversity Increase φ, ψ diversity and accuracy target sequence multiple sequence alignment sequence database MQIFVKTLTGKTITLEV 180° ψ 180° -180° φ homology-free sampling 180° ψ SPEED sampling -180° φ 180° IEIKIRDIYSKTYKFMA IEITCNDRLGKKVRVKC MRLFIRSHLHDQVVISA MKLSVKSPNGRIEIFNE LQFFVRLLDGKSVTLTF IEITLNDRLGKKIRVKC IEIWVNDHLSHRERIKC MDVFLMIRRQKTTIFDA IIVTVNDRLGTKAQIPA MRISVIKLDSTSFDVAV MNVNFRTILGKTYTITV MLLTVRDRSELTFSLQV MQIFVTTPSENVFGLEV MSLTIKF-GAKSIALSL MKYRIRTISNDEAVIEL … ~1000 sequences Uses sequence data base 107 seq’s, growing fast; PDB only 104 structures growing slowly

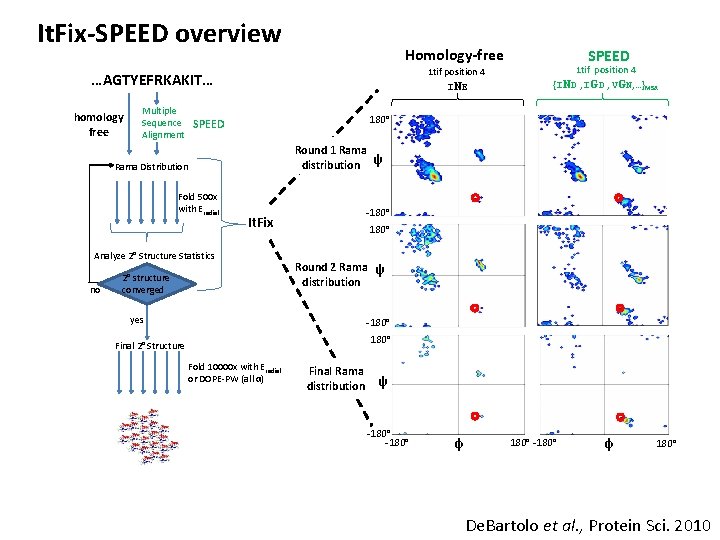
It. Fix-SPEED overview Homology-free 1 tif position 4 INE …AGTYEFRKAKIT… homology free Multiple Sequence Alignment 180° SPEED Round 1 Rama distribution ψ Rama Distribution Fold 500 x with Eradial -180° It. Fix Analyze 2° Structure Statistics no SPEED 1 tif position 4 {IND , IGD , VGN, …}MSA 2° structure converged 180° Round 2 Rama ψ distribution yes -180° Final 2° Structure Fold 10000 x with Eradial or DOPE-PW (all α) Final Rama distribution ψ -180° φ 180° De. Bartolo et al. , Protein Sci. 2010

It. Fix-SPEED overview Homology-free 1 tif position 4 INE …AGTYEFRKAKIT… homology free Multiple Sequence Alignment 180° SPEED Round 1 Rama distribution ψ Rama Distribution Fold 500 x with Eradial -180° It. Fix Analyze 2° Structure Statistics no SPEED 1 tif position 4 {IND , IGD , VGN, …}MSA 180° Round 2 Rama ψ distribution 2° structure converged yes -180° Final 2° Structure Fold 10000 x with Eradial or DOPE-PW (all α) Final Rama distribution ψ cluster Largest cluster Refine 100 X each with DOPE-PW Reject ∆Eradial> 0 -180° φ 180° prediction min<Energy> 100 De. Bartolo et al. , Protein Sci. 2010

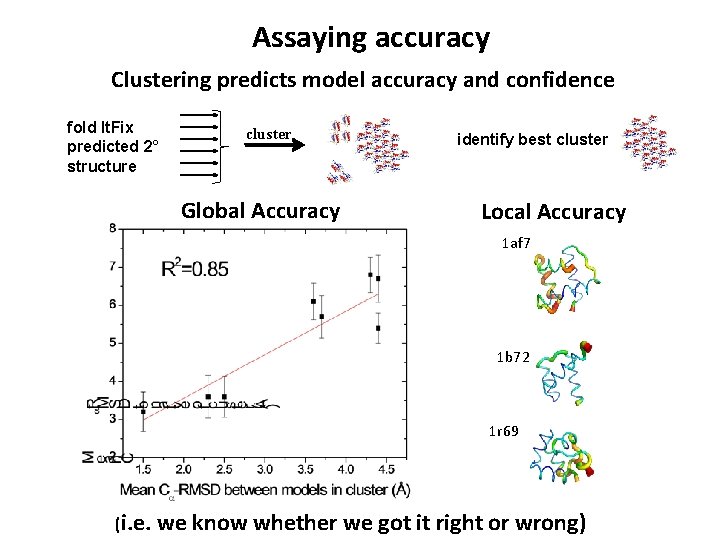
Assaying accuracy Clustering predicts model accuracy and confidence fold It. Fix predicted 2° structure cluster Global Accuracy identify best cluster Local Accuracy 1 af 7 1 b 72 1 r 69 (i. e. we know whether we got it right or wrong)

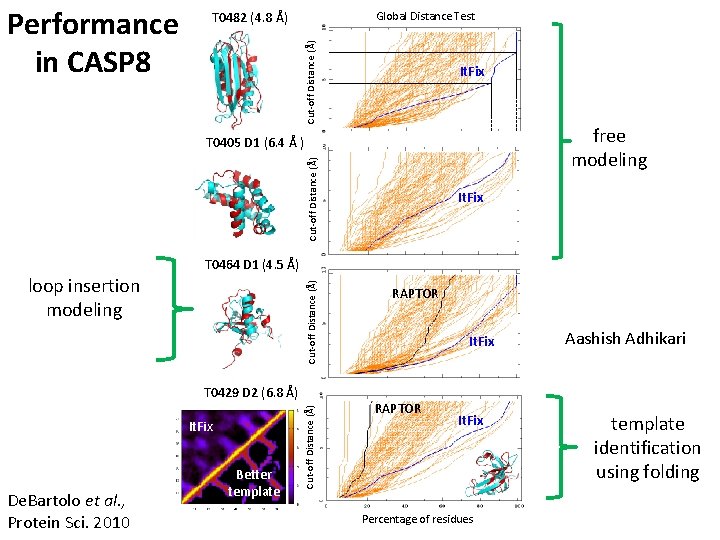
Cut-off Distance (Å) Performance in CASP 8 Global Distance Test T 0482 (4. 8 Å) It. Fix free modeling Cut-off Distance (Å) T 0405 D 1 (6. 4 Å ) It. Fix T 0464 D 1 (4. 5 Å) Cut-off Distance (Å) loop insertion modeling It. Fix De. Bartolo et al. , Protein Sci. 2010 Better template Cut-off Distance (Å) T 0429 D 2 (6. 8 Å) RAPTOR It. Fix Percentage of residues Aashish Adhikari template identification using folding

Part II Conclusions • Adding evolutionary information to It. Fix improves the accuracy of the conformational search • Clustering permits global and local prediction of cluster accuracy and uncertainty • SPEED is successful in the CASP 8 experiment

Protein structure prediction and design PART III PART IV It. Fix: Homology-free structure prediction SPEED: It. Fix enhanced with evolution Future directions in prediction Protein design 1° structure MQIFVKTLTGKTITLEVEPSDTIENVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLSDYNIQKESTLHLVLR

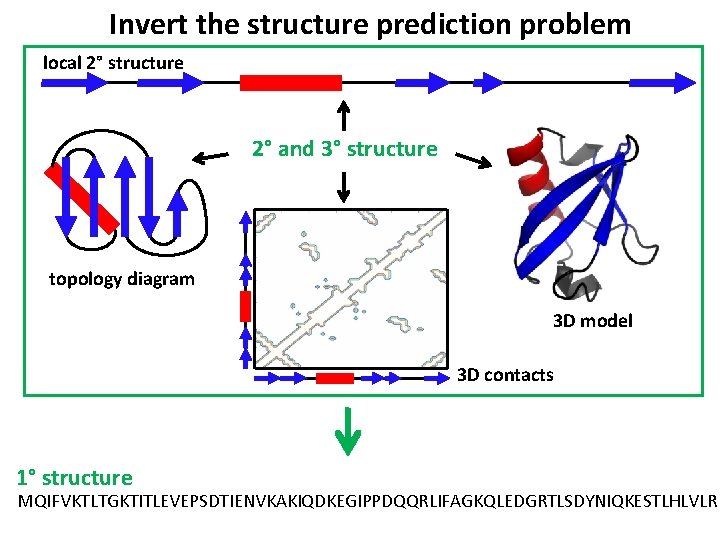
Invert the structure prediction problem local 2° structure 2° and 3° structure topology diagram 3 D model 3 D contacts 1° structure MQIFVKTLTGKTITLEVEPSDTIENVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLSDYNIQKESTLHLVLR

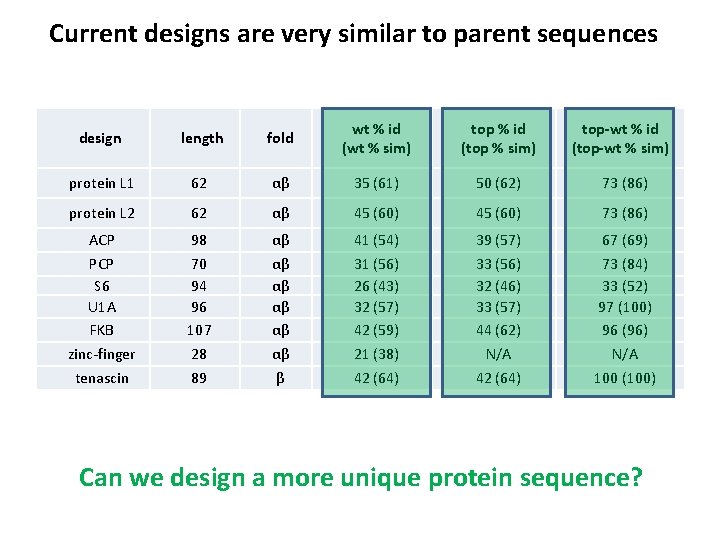
Current designs are very similar to parent sequences design length fold wt % id (wt % sim) top % id (top % sim) top-wt % id (top-wt % sim) protein L 1 62 αβ 35 (61) 50 (62) 73 (86) protein L 2 62 αβ 45 (60) 73 (86) ACP PCP S 6 U 1 A FKB zinc-finger tenascin 98 70 94 96 107 28 89 αβ αβ αβ β 41 (54) 31 (56) 26 (43) 32 (57) 42 (59) 21 (38) 42 (64) 39 (57) 33 (56) 32 (46) 33 (57) 44 (62) N/A 42 (64) 67 (69) 73 (84) 33 (52) 97 (100) 96 (96) N/A 100 (100) Can we design a more unique protein sequence?

Design method 01010111 1 Restrict AA possibilities by burial in native structure for the hydrophobic effect 2 Find best sequences for maximum Rama propensity 3 Monte Carlo search of Statistical Potential DOPE PW energy DOPE-PW GLU-Cβ - GLU-Cβ LEU-Cβ - LEU-Cβ Distance (Å) MKLFVKTP… LTVTIR L IV R E positional sequence library

Hello Jello

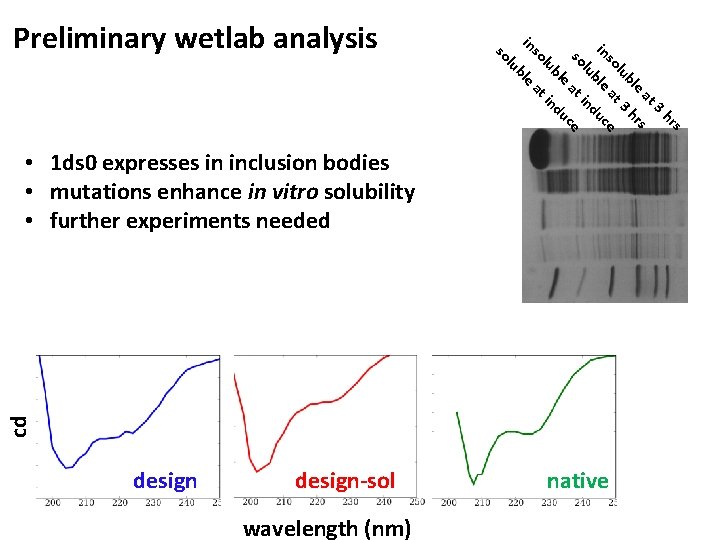
l so s hr 3 at s e r bl 3 h lu at ce so in ble u lu ind s o at ce u le ub ind in lu so Preliminary wetlab analysis e bl at cd • 1 ds 0 expresses in inclusion bodies • mutations enhance in vitro solubility • further experiments needed design-sol wavelength (nm) native

Thesis defense Conclusions • Homology-free structure prediction can provide accurate models by mimicking folding pathways • Adding evolutionary information improves the accuracy of the conformational search • Inverting our homology-free prediction method into a design algorithm aims to generate unique amino acid sequences

Acknowledgements Prof. Tobin Sosnick Prof. Karl Freed Prof. Jinbo Xu Glen Hocky Andres Colubri James Fitzgerald Abhishek Jha Esmael Haddadian James Hinshaw Aashish Adhikari Jouko Virtanen Chloe Antoniou Josiah Zayner Feng Zhao Jian Peng Grzegorz Gawlak Srikanth Aravamuthan Funding: NIH, NSF, Joint Theory Institute

Native Rama probability Enhancement of Ramachandran propensity ψ φ AA Sec. Str position Enhancement in energy and structure prediction • • ∆∆E = -120 (arb. units) 2 X enhancement in native-like models in prediction

1 b 72 1. 6 Å 1 di 2 4. 6 Å 1 r 69 2. 4 Å 1 1 af 7 2. 7 Å Round 0 Round 1 Round 2 Round 3 Round 4 Round 3 Round 5 Round 4 Round 6 Round 4 Round 7 Round 6 Round 8 Round 6 10 10 1 0 10 0 Secondary Structure frequency 10 Round 0 residue index

SPEED increases the native Rama probability native Rama regions 180 2 ψ -180 Native basin probability 1 1 b 72 φ % positions with PNative > 0. 25 3 SPEED reduces cases SPEED whereimproves native φ, native ψ hasφ, aψ probability sequence very lowacross probability 4 180 2° structure by position PDB of target Amino acid by id position

Radial energy terms enforce productive chain collapse (global terms) Rg-Cα: Root-squared distance of Cα from CM. Compactness of model Rg-phil Rg-phob CMCα Rg-Cα Cα Cβ Ru-Cα: Root-meansquared deviation of Cα from CM. Enforces a spherical model Rg-phob/Rg-phil (burial ratio): best packing of hydrophobic residues

Eliminating the fixing round 0 thresholds from It. Fix round 1 180 (e. g. pos. 67) MQIFVKT…STLHLVLR Rama distribution 0 -180 180 fold 2000 X round 2 Rama distribution 0 -180 fold 2000 X round 3 Rama distribution 0 -180 0 180

An evolution-enhanced energy function DOPE-PW-SPEED 10 WT: ILE Homologs: polar DOPE-PW-SPEED WT: Ala Homologs: polar energy 8 6 4 2 0 0. 0 10 PHE 4 THR 14 energy 8 5. 0 10. 0 15. 0 20. 0 25. 0 30. 0 distance (Å) DOPE-PW-SPEED 6 4 2 0 0. 0 2. 0 4. 0 6. 0 8. 0 10. 0 12. 0 14. 0 16. 0 distance (Å)