Network Modeling through LASSO Regression Farrokh Alemi Ph

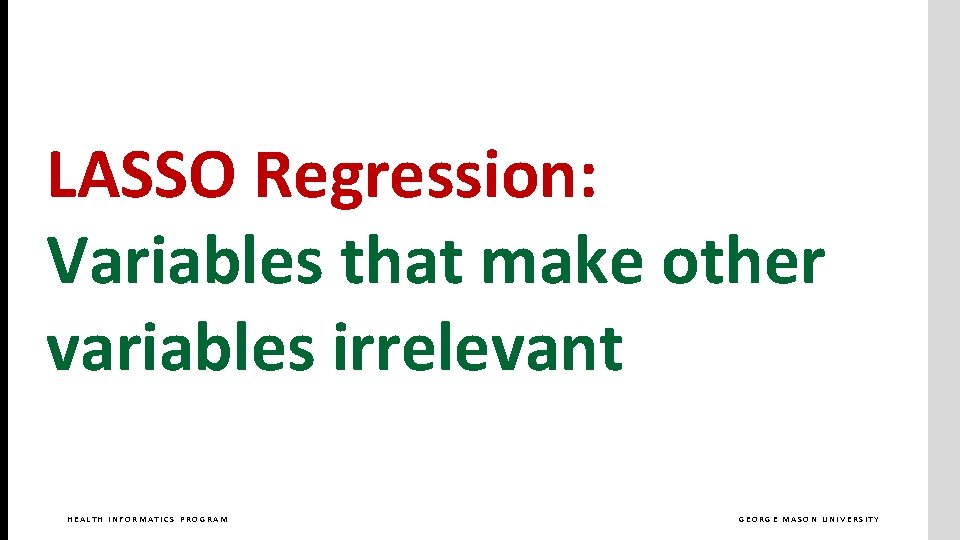








- Slides: 30

Network Modeling through LASSO Regression Farrokh Alemi, Ph. D. HEALTH INFORMATICS PROGRAM HI. GMU. EDU
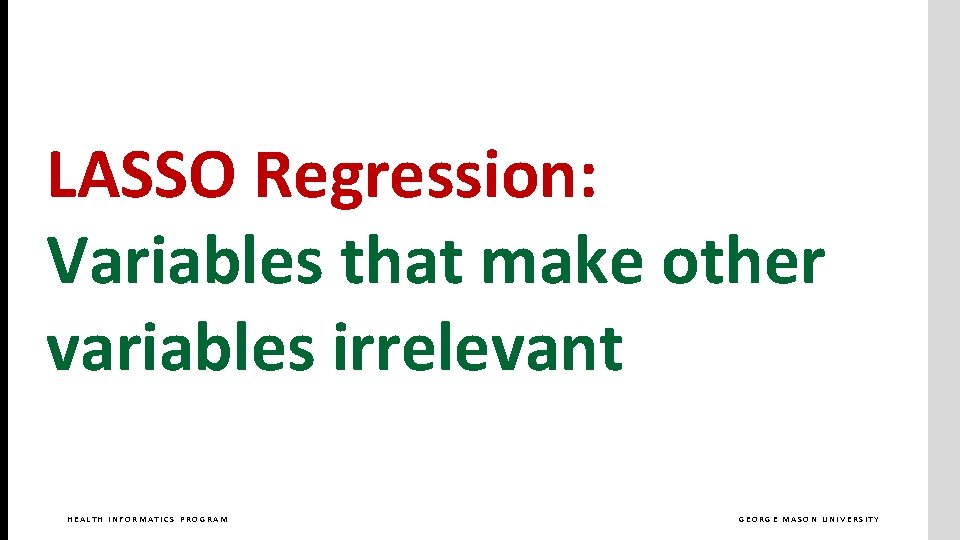
LASSO Regression: Variables that make other variables irrelevant HEALTH INFORMATICS PROGRAM GEORGE MASON UNIVERSITY

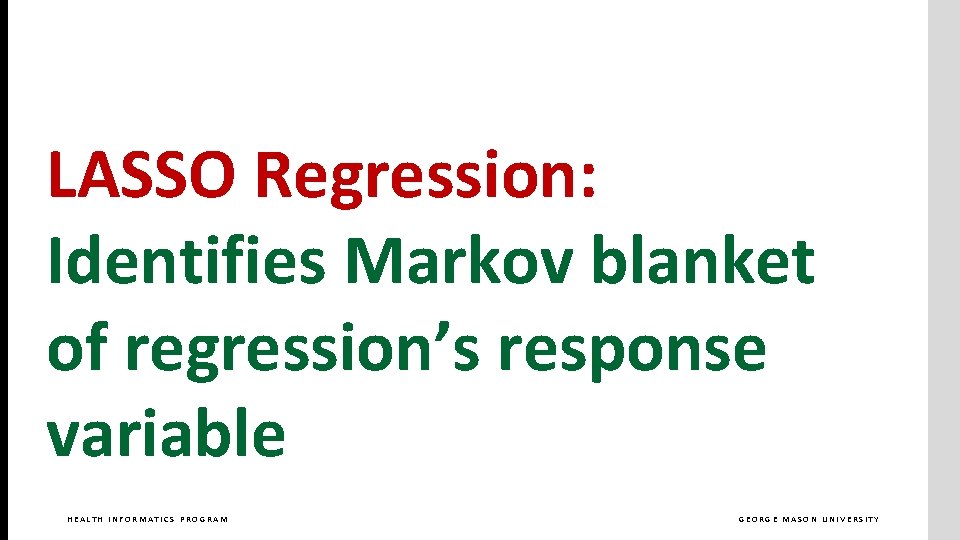
LASSO Regression: Identifies Markov blanket of regression’s response variable HEALTH INFORMATICS PROGRAM GEORGE MASON UNIVERSITY

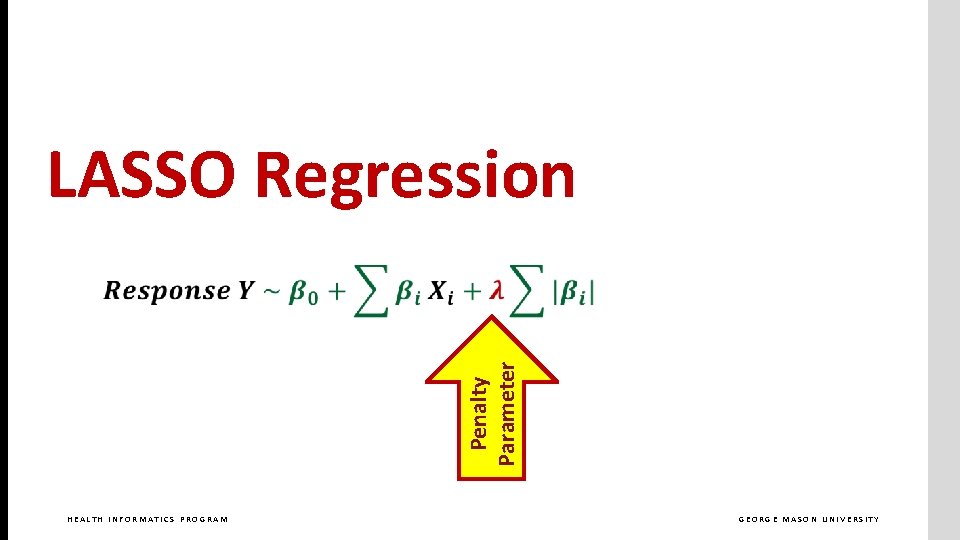
Penalty Parameter LASSO Regression HEALTH INFORMATICS PROGRAM GEORGE MASON UNIVERSITY

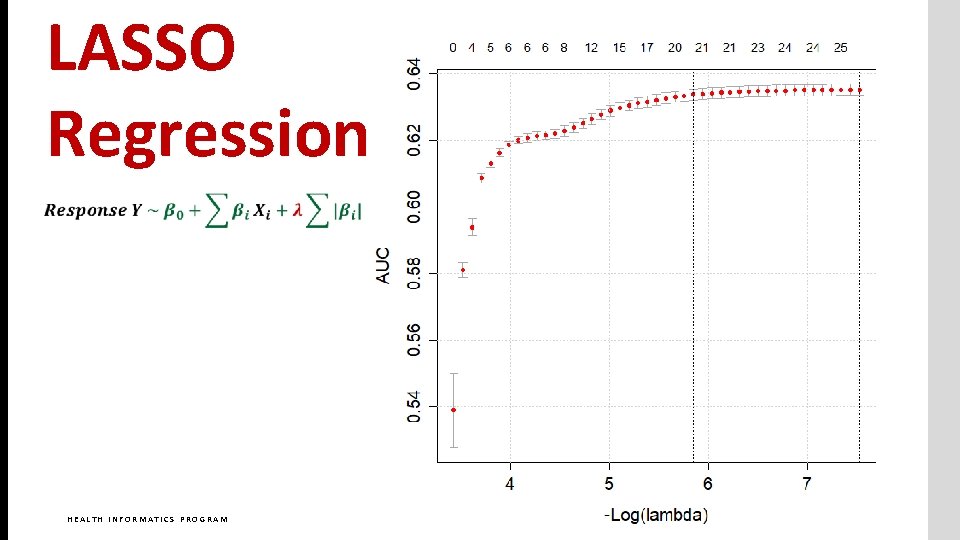
LASSO Regression HEALTH INFORMATICS PROGRAM GEORGE MASON UNIVERSITY

Causal Networks: Parents in Markov blankets HEALTH INFORMATICS PROGRAM GEORGE MASON UNIVERSITY

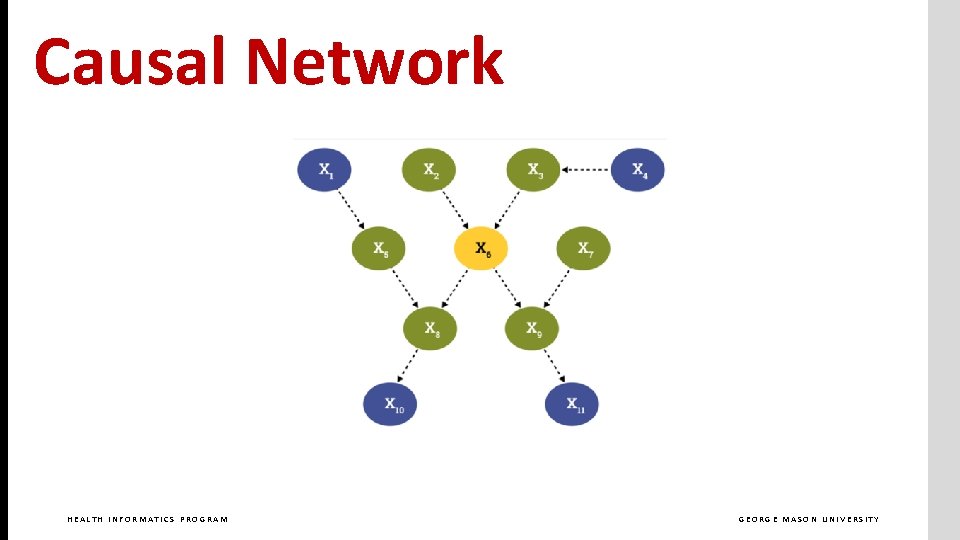
Causal Network HEALTH INFORMATICS PROGRAM GEORGE MASON UNIVERSITY

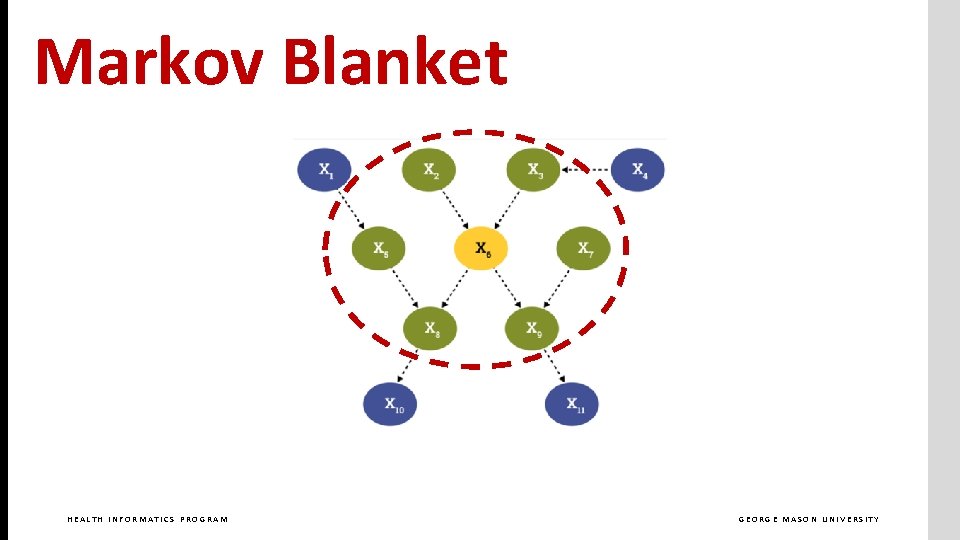
Markov Blanket HEALTH INFORMATICS PROGRAM GEORGE MASON UNIVERSITY

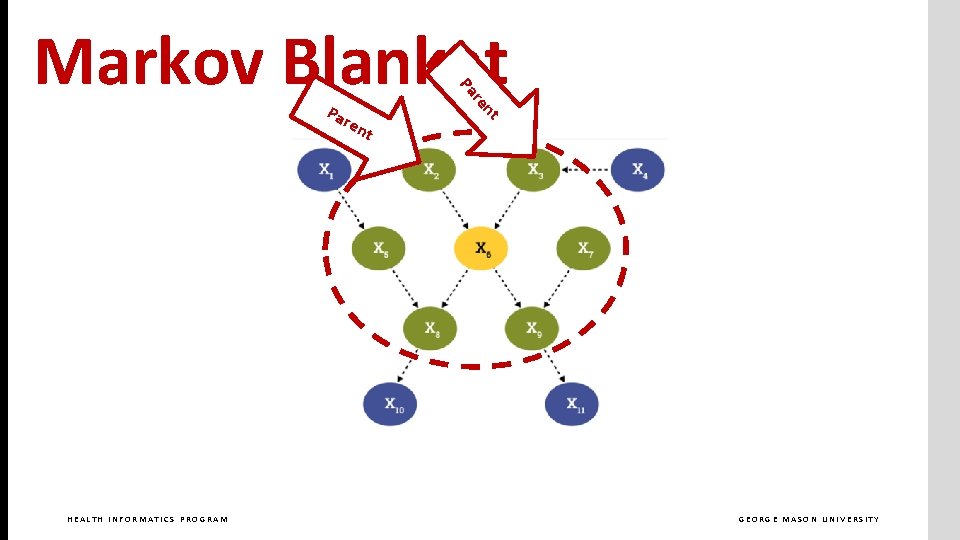
ren HEALTH INFORMATICS PROGRAM t nt Pa re Pa Markov Blanket GEORGE MASON UNIVERSITY

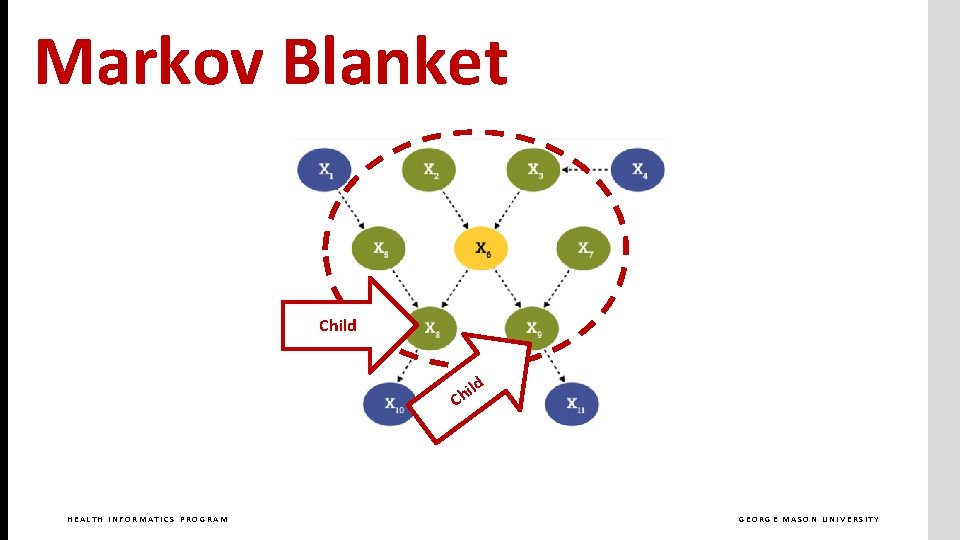
Markov Blanket Child d il Ch HEALTH INFORMATICS PROGRAM GEORGE MASON UNIVERSITY

Markov Blanket Co-parents ts n e r a -p Co HEALTH INFORMATICS PROGRAM GEORGE MASON UNIVERSITY

LASSO Regression on preceding variables identifies parents in Markov blanket HEALTH INFORMATICS PROGRAM GEORGE MASON UNIVERSITY

Order of Occurrence: 1. 2. 3. By Definition By average time of occurrence By time of measurement HEALTH INFORMATICS PROGRAM GEORGE MASON UNIVERSITY

An Example HEALTH INFORMATICS PROGRAM GEORGE MASON UNIVERSITY

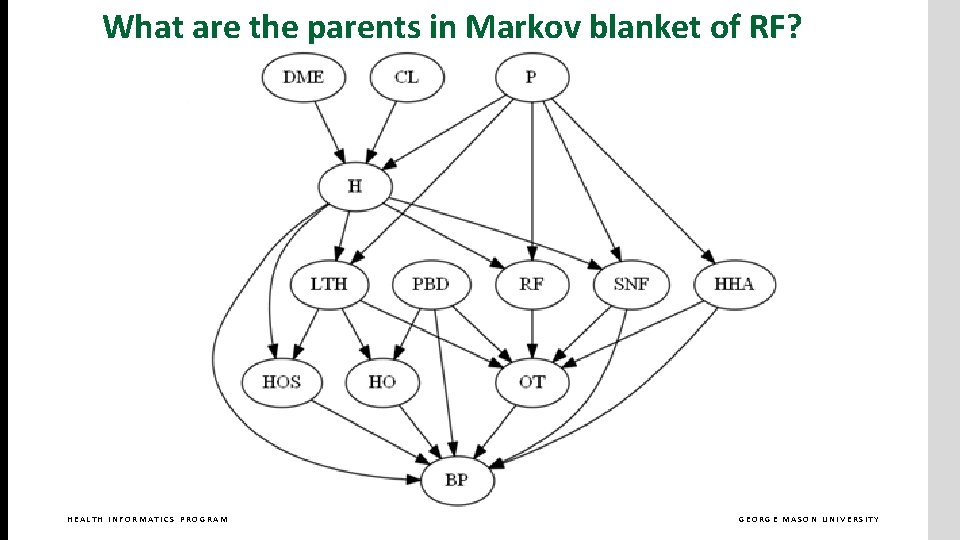
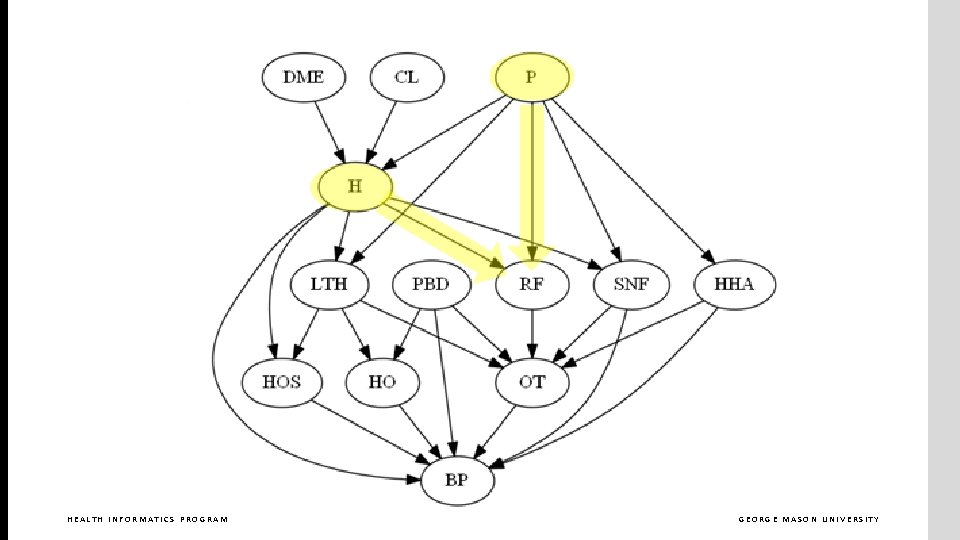
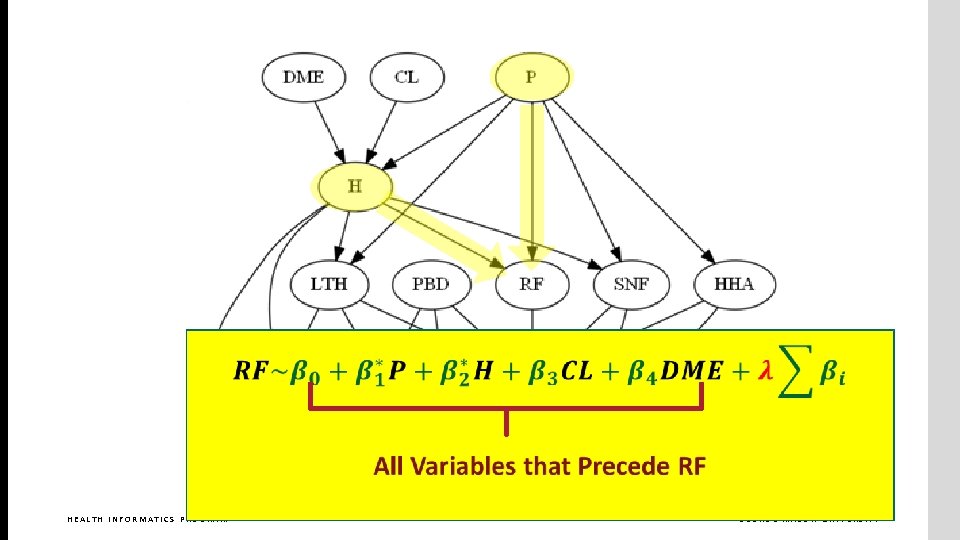
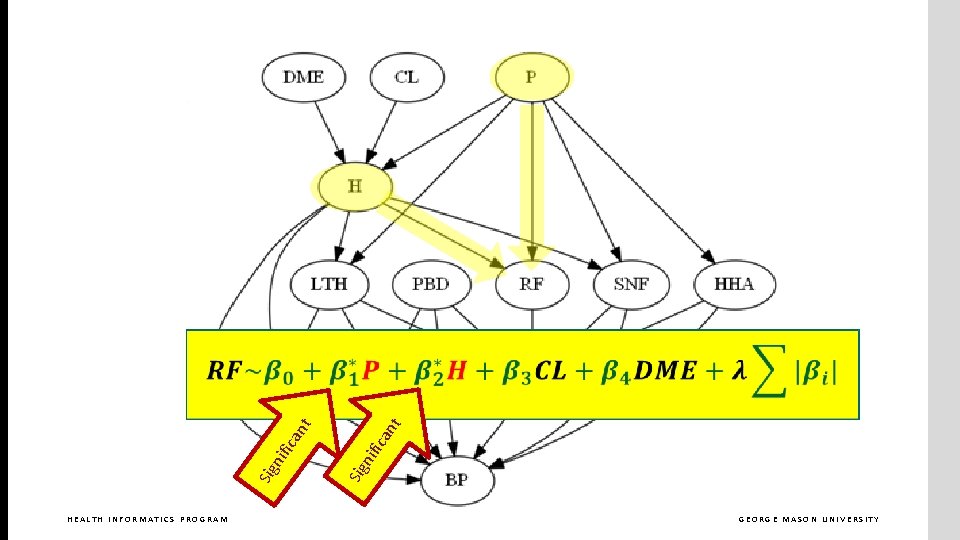
What are the parents in Markov blanket of RF? HEALTH INFORMATICS PROGRAM GEORGE MASON UNIVERSITY

HEALTH INFORMATICS PROGRAM GEORGE MASON UNIVERSITY

HEALTH INFORMATICS PROGRAM GEORGE MASON UNIVERSITY

nt ica nif Sig HEALTH INFORMATICS PROGRAM GEORGE MASON UNIVERSITY

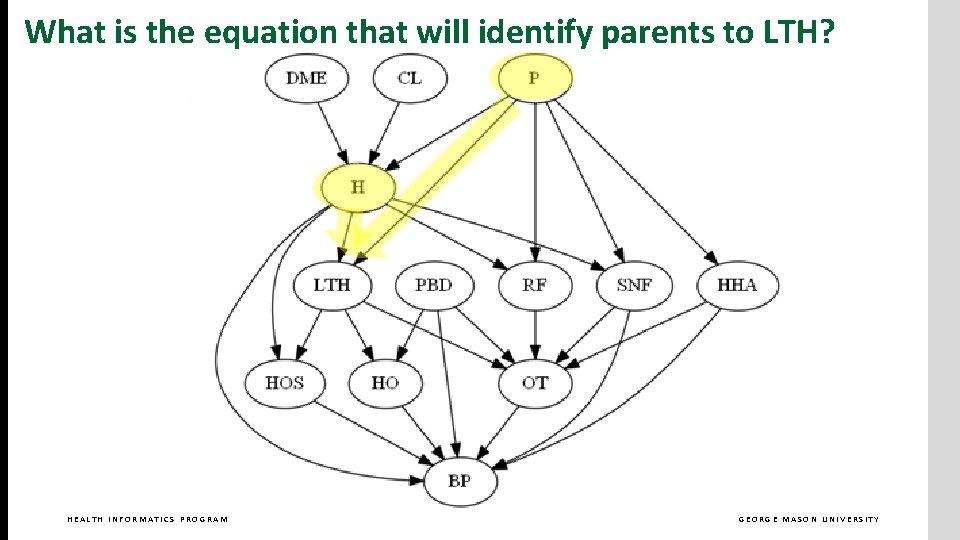
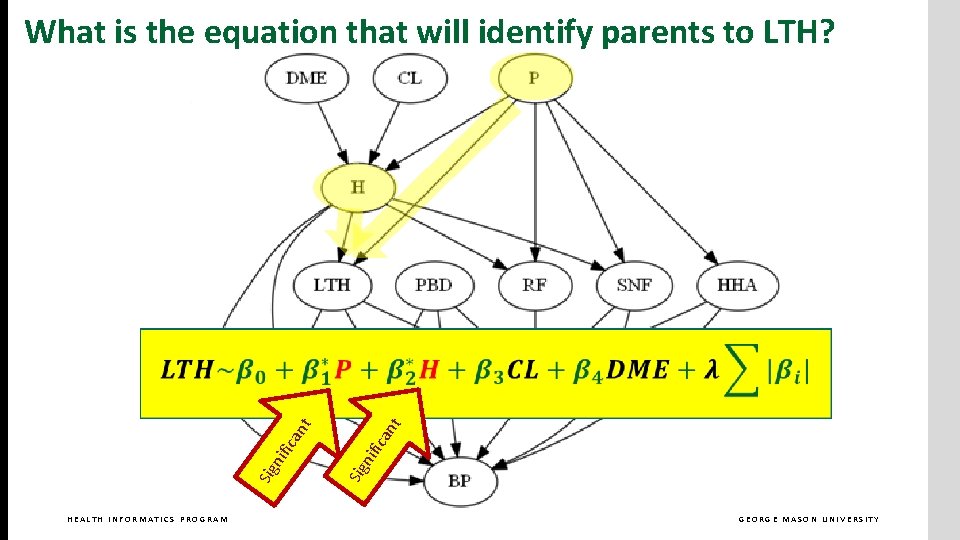
What is the equation that will identify parents to LTH? HEALTH INFORMATICS PROGRAM GEORGE MASON UNIVERSITY

HEALTH INFORMATICS PROGRAM nt ica nif Sig nif ica nt What is the equation that will identify parents to LTH? GEORGE MASON UNIVERSITY

Repeated LASSO Regression Response Variable: Each Variable in Analysis Independent Variables: All Variables Preceding Response HEALTH INFORMATICS PROGRAM GEORGE MASON UNIVERSITY

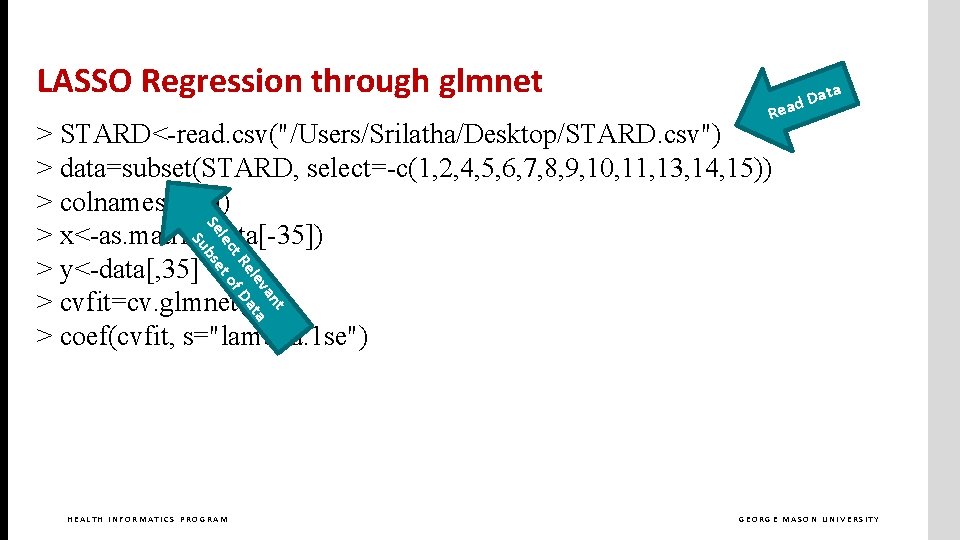
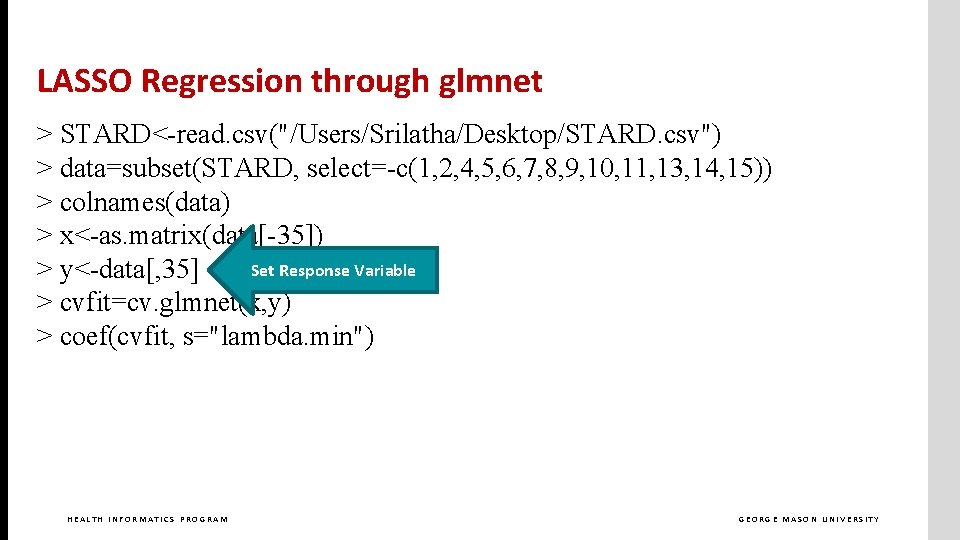
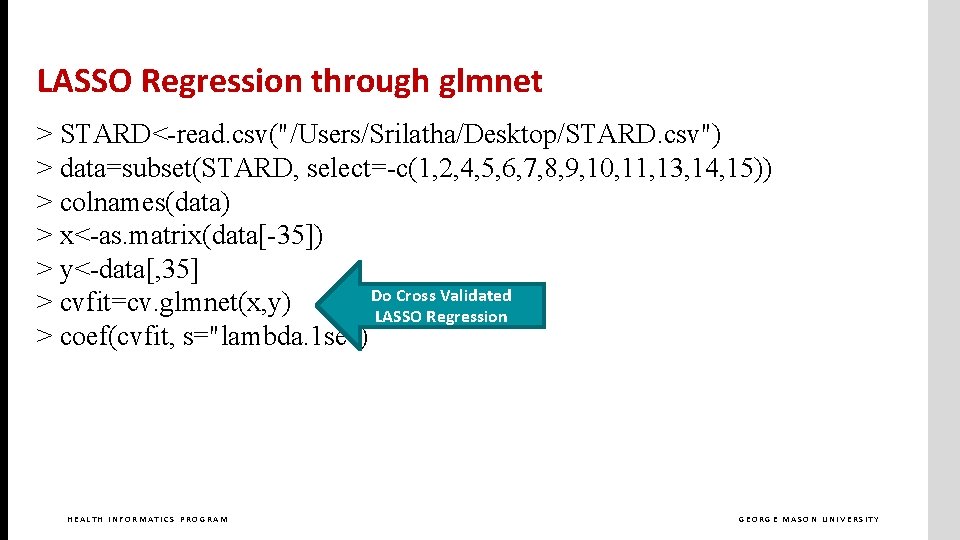
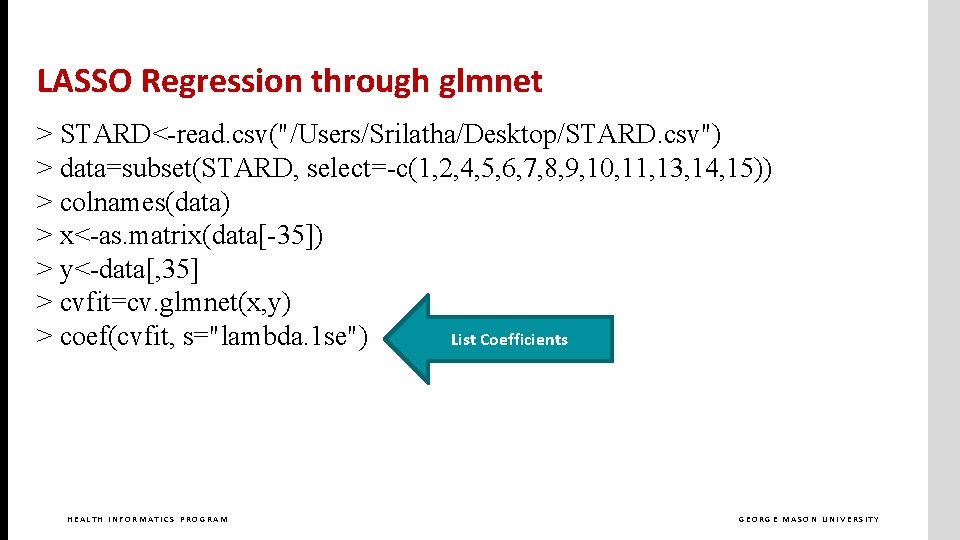
LASSO Regression through glmnet > STARD<-read. csv("/Users/Srilatha/Desktop/STARD. csv") > data=subset(STARD, select=-c(1, 2, 4, 5, 6, 7, 8, 9, 10, 11, 13, 14, 15)) > colnames(data) > x<-as. matrix(data[-35]) > y<-data[, 35] > cvfit=cv. glmnet(x, y) > coef(cvfit, s="lambda. 1 se") HEALTH INFORMATICS PROGRAM GEORGE MASON UNIVERSITY

LASSO Regression through glmnet ata D d ea R nt va a ele at t. R f. D lec t o Se bse Su > STARD<-read. csv("/Users/Srilatha/Desktop/STARD. csv") > data=subset(STARD, select=-c(1, 2, 4, 5, 6, 7, 8, 9, 10, 11, 13, 14, 15)) > colnames(data) > x<-as. matrix(data[-35]) > y<-data[, 35] > cvfit=cv. glmnet(x, y) > coef(cvfit, s="lambda. 1 se") HEALTH INFORMATICS PROGRAM GEORGE MASON UNIVERSITY

LASSO Regression through glmnet > STARD<-read. csv("/Users/Srilatha/Desktop/STARD. csv") > data=subset(STARD, select=-c(1, 2, 4, 5, 6, 7, 8, 9, 10, 11, 13, 14, 15)) > colnames(data) Exclude response from > x<-as. matrix(data[-35]) independent variables > y<-data[, 35] > cvfit=cv. glmnet(x, y) > coef(cvfit, s="lambda. 1 se") HEALTH INFORMATICS PROGRAM GEORGE MASON UNIVERSITY

LASSO Regression through glmnet > STARD<-read. csv("/Users/Srilatha/Desktop/STARD. csv") > data=subset(STARD, select=-c(1, 2, 4, 5, 6, 7, 8, 9, 10, 11, 13, 14, 15)) > colnames(data) > x<-as. matrix(data[-35]) Set Response Variable > y<-data[, 35] > cvfit=cv. glmnet(x, y) > coef(cvfit, s="lambda. min") HEALTH INFORMATICS PROGRAM GEORGE MASON UNIVERSITY

LASSO Regression through glmnet > STARD<-read. csv("/Users/Srilatha/Desktop/STARD. csv") > data=subset(STARD, select=-c(1, 2, 4, 5, 6, 7, 8, 9, 10, 11, 13, 14, 15)) > colnames(data) > x<-as. matrix(data[-35]) > y<-data[, 35] Do Cross Validated > cvfit=cv. glmnet(x, y) LASSO Regression > coef(cvfit, s="lambda. 1 se") HEALTH INFORMATICS PROGRAM GEORGE MASON UNIVERSITY

LASSO Regression through glmnet > STARD<-read. csv("/Users/Srilatha/Desktop/STARD. csv") > data=subset(STARD, select=-c(1, 2, 4, 5, 6, 7, 8, 9, 10, 11, 13, 14, 15)) > colnames(data) > x<-as. matrix(data[-35]) > y<-data[, 35] > cvfit=cv. glmnet(x, y) > coef(cvfit, s="lambda. 1 se") List Coefficients HEALTH INFORMATICS PROGRAM GEORGE MASON UNIVERSITY

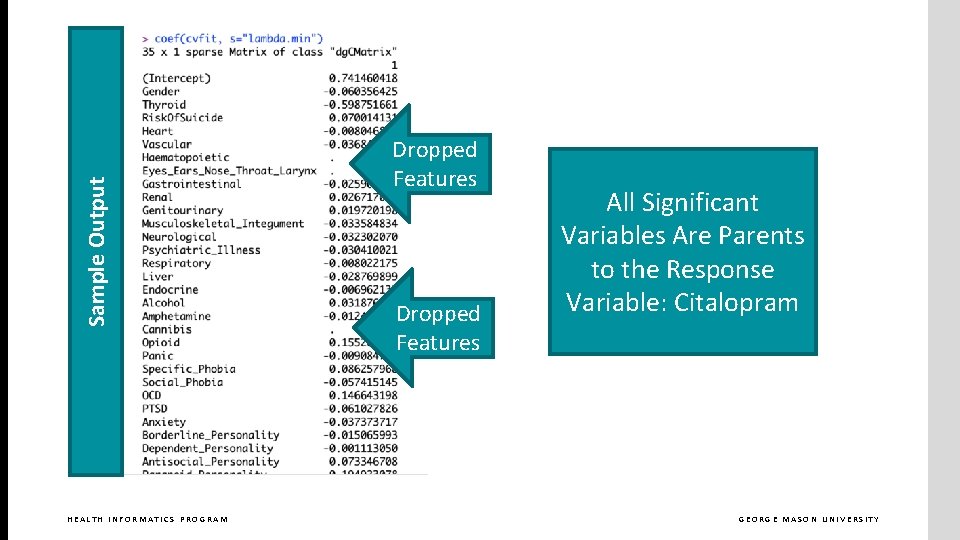
Sample Output HEALTH INFORMATICS PROGRAM Dropped Features All Significant Variables Are Parents to the Response Variable: Citalopram GEORGE MASON UNIVERSITY

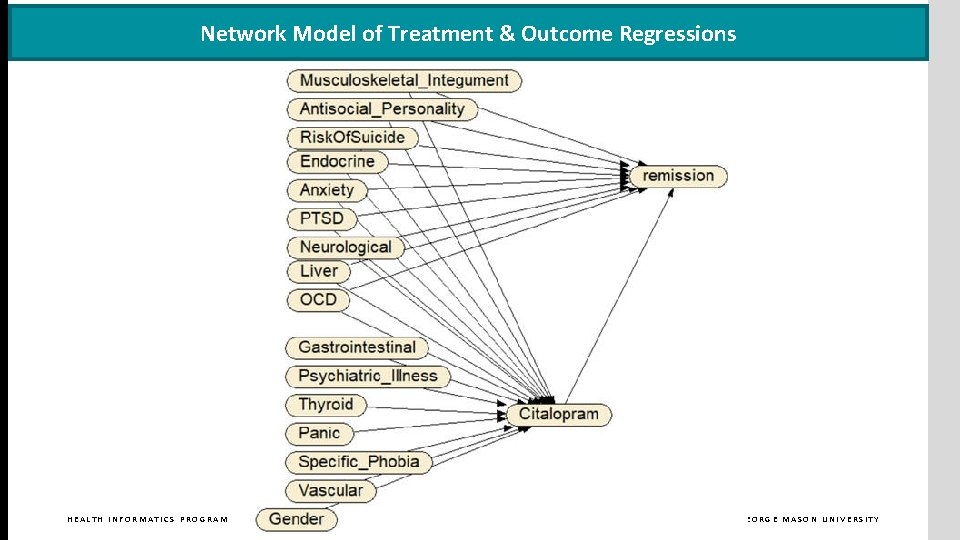
Network Model of Treatment & Outcome Regressions HEALTH INFORMATICS PROGRAM GEORGE MASON UNIVERSITY

REPEATED LASSO REGRESSION IDENTIFIES ENTIRE NETWORK STRUCTURE