National Taiwan University Department of Computer Science and
























- Slides: 54

National Taiwan University Department of Computer Science and Information Engineering Introduction to SNP and Haplotype Analysis Yao-Ting Huang Kun-Mao Chao Algorithms and Computational Biology Lab, Department of Computer Science & Information Engineering, National Taiwan University, Taiwan. 3/11/2021

National Taiwan University Department of Computer Science and Information Engineering Genetic Variations n The genetic variations in DNA sequences (e. g. , insertions, deletions, and mutations) have a major impact on genetic diseases and phenotypic differences. All humans share 99% the same DNA sequence. u The genetic variations in the coding region may change the codon of an amino acid and alters the amino acid sequence. u 2

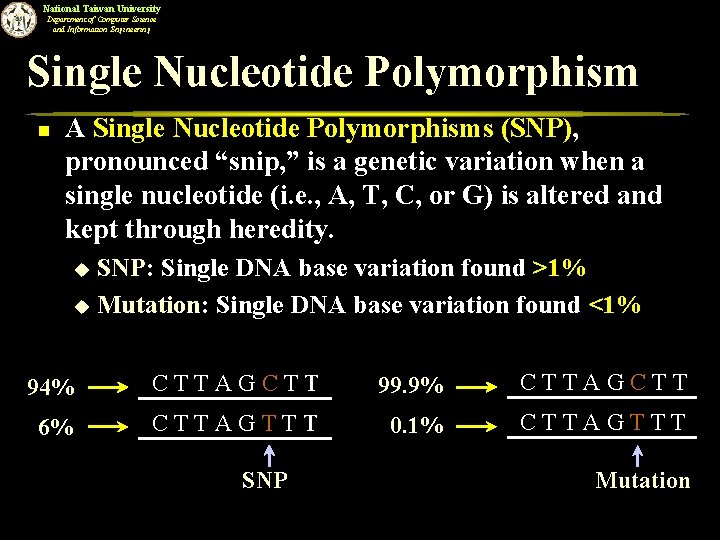
National Taiwan University Department of Computer Science and Information Engineering Single Nucleotide Polymorphism n A Single Nucleotide Polymorphisms (SNP), pronounced “snip, ” is a genetic variation when a single nucleotide (i. e. , A, T, C, or G) is altered and kept through heredity. SNP: Single DNA base variation found >1% u Mutation: Single DNA base variation found <1% u 94% CTTAGCTT 99. 9% CTTAGCTT 6% CTTAGTTT 0. 1% CTTAGTTT SNP Mutation

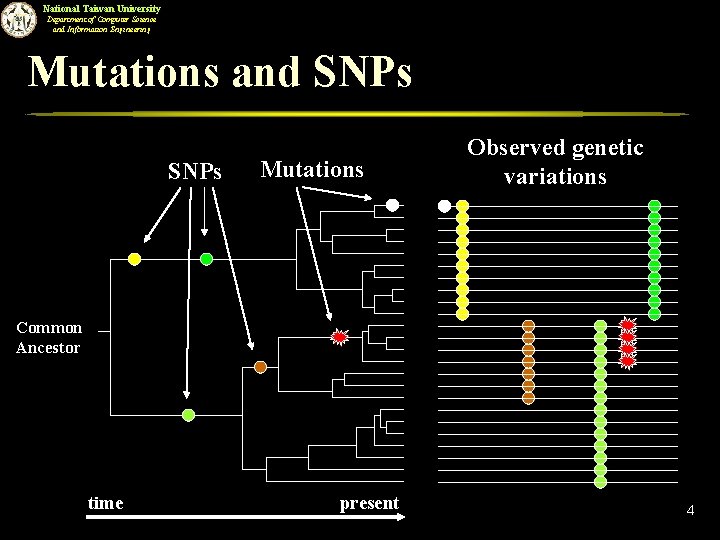
National Taiwan University Department of Computer Science and Information Engineering Mutations and SNPs Mutations Observed genetic variations Common Ancestor time present 4

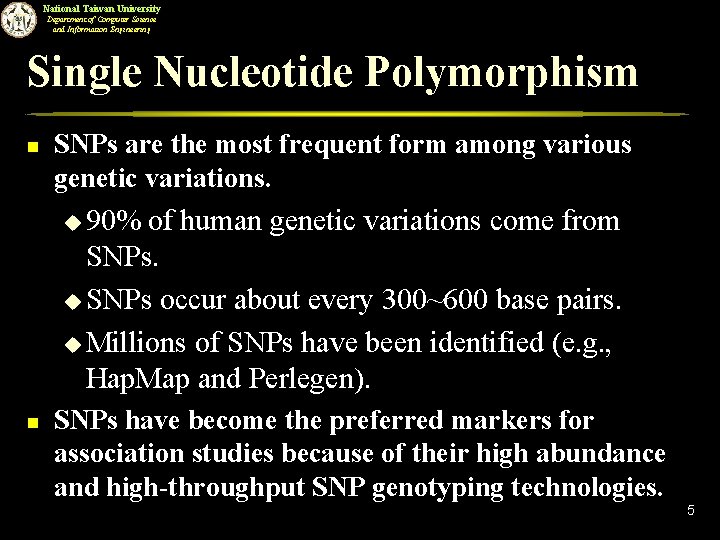
National Taiwan University Department of Computer Science and Information Engineering Single Nucleotide Polymorphism n SNPs are the most frequent form among various genetic variations. u 90% of human genetic variations come from SNPs. u SNPs occur about every 300~600 base pairs. u Millions of SNPs have been identified (e. g. , Hap. Map and Perlegen). n SNPs have become the preferred markers for association studies because of their high abundance and high-throughput SNP genotyping technologies. 5

National Taiwan University Department of Computer Science and Information Engineering Single Nucleotide Polymorphism n A SNP is usually assumed to be a binary variable. The probability of repeat mutation at the same SNP locus is quite small. u The tri-allele cases are usually considered to be the effect of genotyping errors. u n The nucleotide on a SNP locus is called a major allele (if allele frequency > 50%), or u a minor allele (if allele frequency < 50%). u 94% ACTTAGCTT T: Major allele 6% ACTTAGCTC C: Minor allele

National Taiwan University Department of Computer Science and Information Engineering Haplotypes n A haplotype stands for a set of linked SNPs on the same chromosome. u -AAChaplotype TTTGC can considered T C-be simply. Haplotype 1 as. Ca binary string since each SNP is binary. C A T -A C T T A G C T T- Haplotype 2 -A A T T T G C T C- Haplotype 3 A SNP 1 SNP 2 SNP 3 T C SNP 1 SNP 2 SNP 3 7

National Taiwan University Department of Computer Science and Information Engineering Genotypes n The use of haplotype information has been limited because the human genome is a diploid. u In large sequencing projects, genotypes instead of haplotypes are collected due to cost consideration. A G C T SNP 1 SNP 2 A C G A T T SNP 1 SNP 2 Genotype data C G SNP 1 SNP 2 A C T G SNP 1 SNP 2 Haplotype data 8

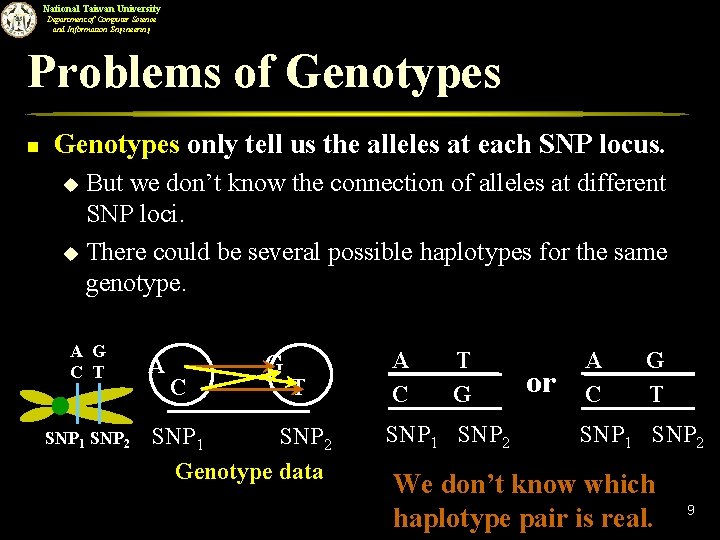
National Taiwan University Department of Computer Science and Information Engineering Problems of Genotypes n Genotypes only tell us the alleles at each SNP locus. But we don’t know the connection of alleles at different SNP loci. u There could be several possible haplotypes for the same genotype. u A G C T SNP 1 SNP 2 A C G T SNP 1 SNP 2 Genotype data A C T G SNP 1 SNP 2 or A C G T SNP 1 SNP 2 We don’t know which haplotype pair is real. 9

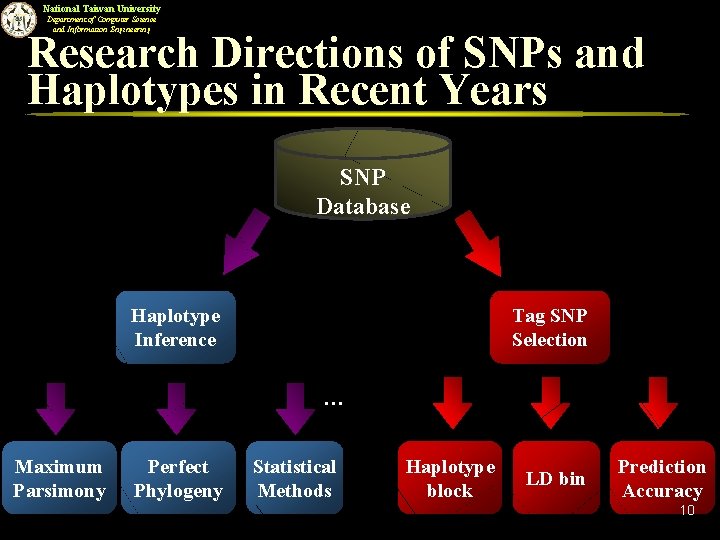
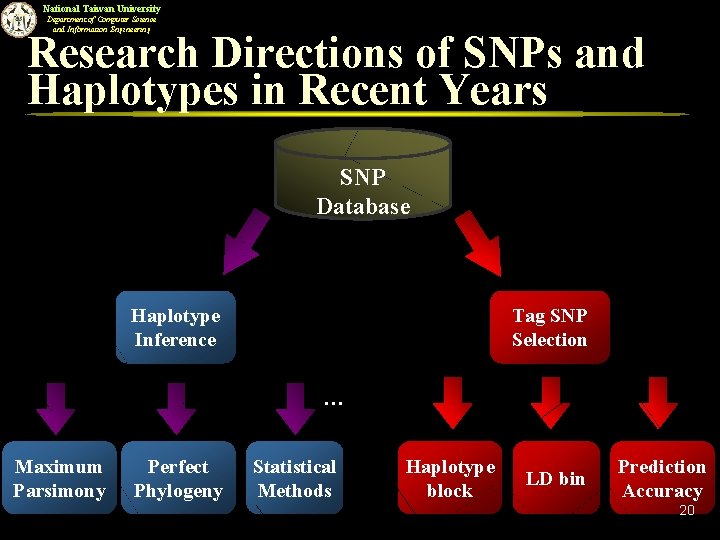
National Taiwan University Department of Computer Science and Information Engineering Research Directions of SNPs and Haplotypes in Recent Years SNP Database Haplotype Inference Tag SNP Selection … Maximum Parsimony Perfect Phylogeny Statistical Methods Haplotype block LD bin Prediction Accuracy 10

National Taiwan University Department of Computer Science and Information Engineering Haplotype Inference n The problem of inferring the haplotypes from a set of genotypes is called haplotype inference. u n This problem is already known to be not only NP-hard but also APX-hard. Most combinatorial methods consider the maximum parsimony model to solve this problem. This model assumes that the real haplotypes in natural population is rare. u The solution of this problem is a minimum set of haplotypes that can explain the given genotypes. u 11

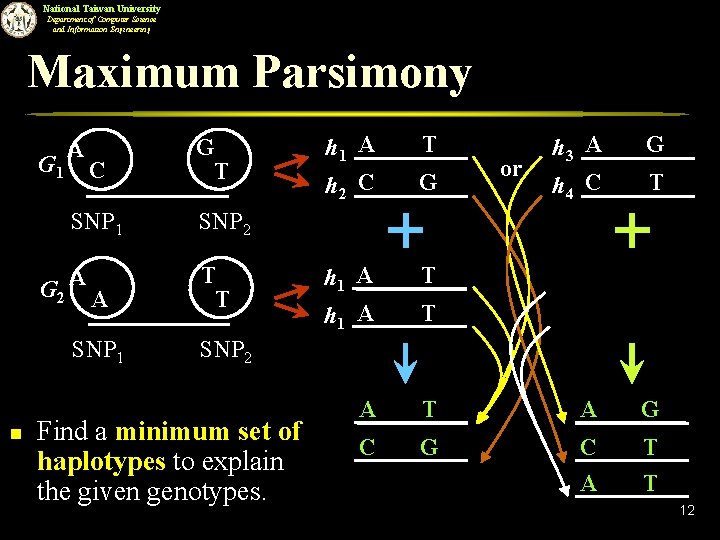
National Taiwan University Department of Computer Science and Information Engineering Maximum Parsimony G 1 A C SNP 1 G 2 A A SNP 1 n G T h 1 A h 2 C T G or h 3 A h 4 C G T SNP 2 T T h 1 A T A G C T A T T SNP 2 Find a minimum set of haplotypes to explain the given genotypes. 12

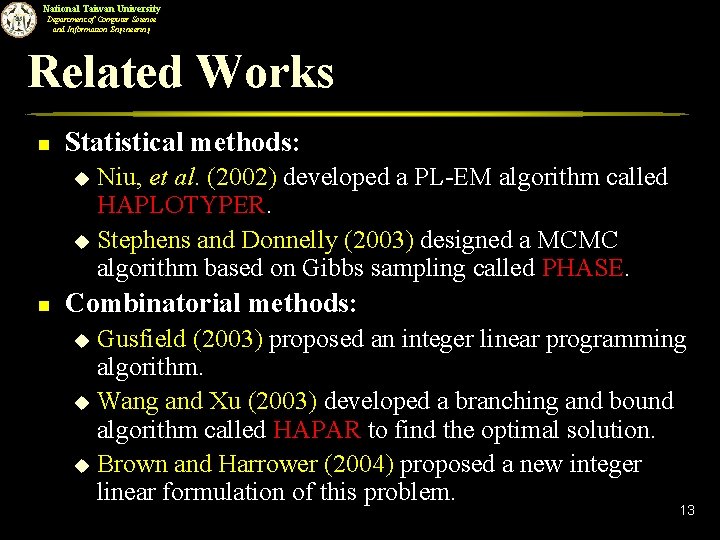
National Taiwan University Department of Computer Science and Information Engineering Related Works n Statistical methods: Niu, et al. (2002) developed a PL-EM algorithm called HAPLOTYPER. u Stephens and Donnelly (2003) designed a MCMC algorithm based on Gibbs sampling called PHASE. u n Combinatorial methods: Gusfield (2003) proposed an integer linear programming algorithm. u Wang and Xu (2003) developed a branching and bound algorithm called HAPAR to find the optimal solution. u Brown and Harrower (2004) proposed a new integer linear formulation of this problem. u 13

National Taiwan University Department of Computer Science and Information Engineering Our Results n n We formulated this problem as an integer quadratic programming (IQP) problem. We proposed an iterative semidefinite programming (SDP) relaxation algorithm to solve the IQP problem. u n This algorithm finds a solution of O(log n) approximation. We implemented this algorithm in Mat. Lab and compared with existing methods. u Huang, Y. -T. , Chao, K. -M. , and Chen, T. , 2005, “An Approximation Algorithm for Haplotype Inference by Maximum Parsimony, ” Journal of Computational Biology, 12: 1261 -1274. 14

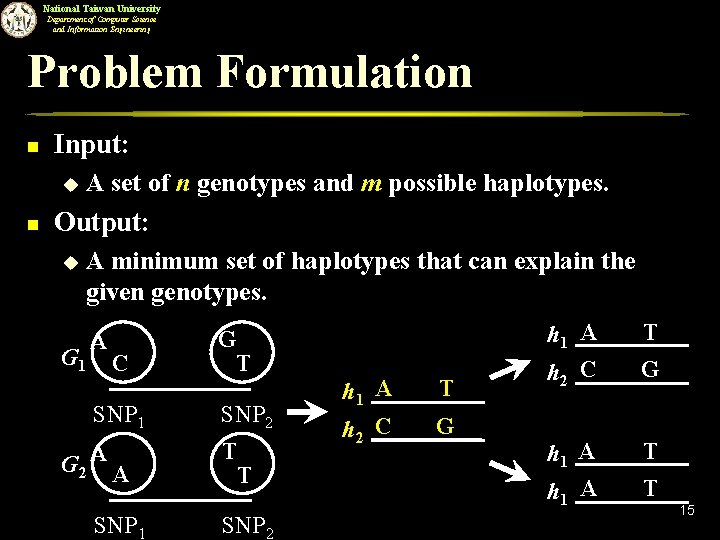
National Taiwan University Department of Computer Science and Information Engineering Problem Formulation n Input: u n A set of n genotypes and m possible haplotypes. Output: u A minimum set of haplotypes that can explain the given genotypes. G 1 A C SNP 1 G 2 A A SNP 1 G T SNP 2 T T SNP 2 h 1 A h 2 C T G h 1 A h 2 C h 1 A T G T T 15

National Taiwan University Department of Computer Science and Information Engineering Integer Quadratic Programming (IQP) n Define xi as an integer variable with values 1 or -1. xi = 1 if the i-th haplotype is selected. u xi = -1 if the i-th haplotype is not selected. u n Minimizing the number of selected haplotypes is to minimize the following integer quadratic function: 16

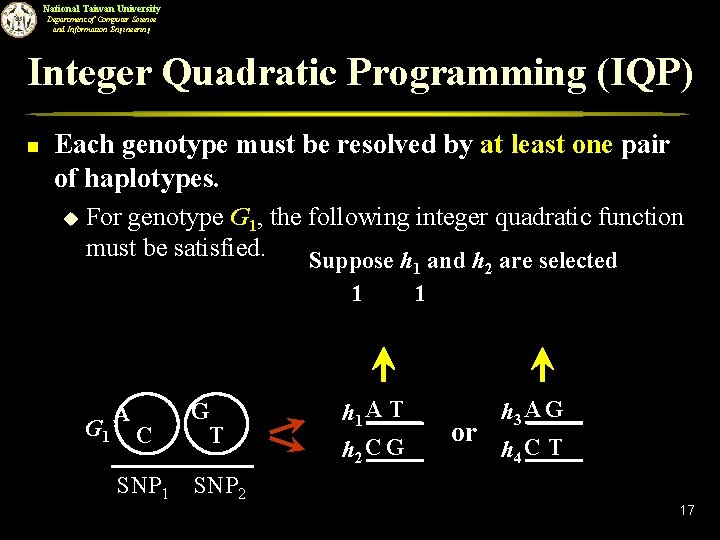
National Taiwan University Department of Computer Science and Information Engineering Integer Quadratic Programming (IQP) n Each genotype must be resolved by at least one pair of haplotypes. u For genotype G 1, the following integer quadratic function must be satisfied. Suppose h and h are selected 1 1 G 1 A C SNP 1 G T SNP 2 h 1 A T h 2 C G 2 1 or h 3 A G h 4 C T 17

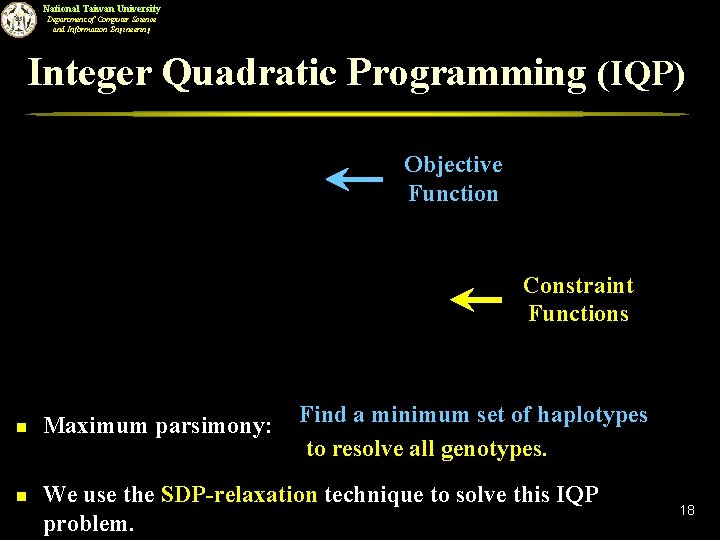
National Taiwan University Department of Computer Science and Information Engineering Integer Quadratic Programming (IQP) Objective Function Constraint Functions n n Maximum parsimony: Find a minimum set of haplotypes to resolve all genotypes. We use the SDP-relaxation technique to solve this IQP problem. 18

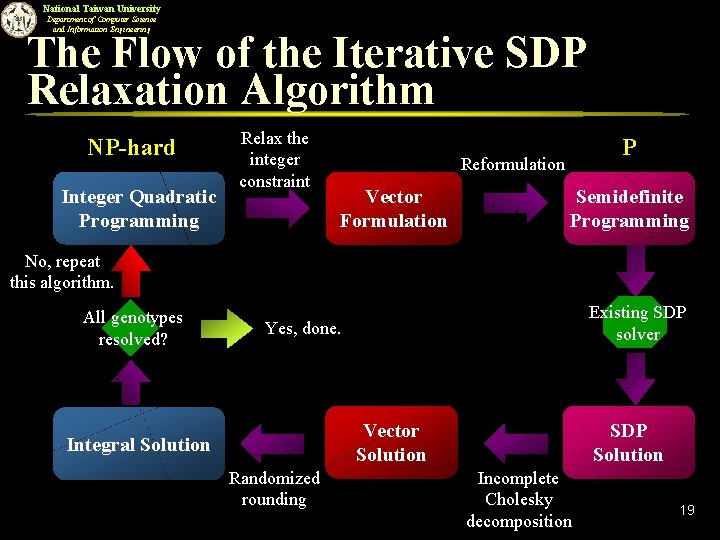
National Taiwan University Department of Computer Science and Information Engineering The Flow of the Iterative SDP Relaxation Algorithm NP-hard Integer Quadratic Programming Relax the integer constraint P Reformulation Vector Formulation Semidefinite Programming No, repeat this algorithm. All genotypes resolved? Existing SDP solver Yes, done. Vector Solution Integral Solution Randomized rounding SDP Solution Incomplete Cholesky decomposition 19

National Taiwan University Department of Computer Science and Information Engineering Research Directions of SNPs and Haplotypes in Recent Years SNP Database Haplotype Inference Tag SNP Selection … Maximum Parsimony Perfect Phylogeny Statistical Methods Haplotype block LD bin Prediction Accuracy 20

National Taiwan University Department of Computer Science and Information Engineering Problems of Using SNPs for Association Studies n The number of SNPs is still too large to be used for association studies. There are millions of SNPs in a human body. u To reduce the SNP genotyping cost, we wish to use as few SNPs as possible for association studies. u n Tag SNPs are a small subset of SNPs that is sufficient for performing association studies without losing the power of using all SNPs. There are many definitions of tag SNPs. u We will first study one definition of tag SNPs based on haplotype blocks model. u 21

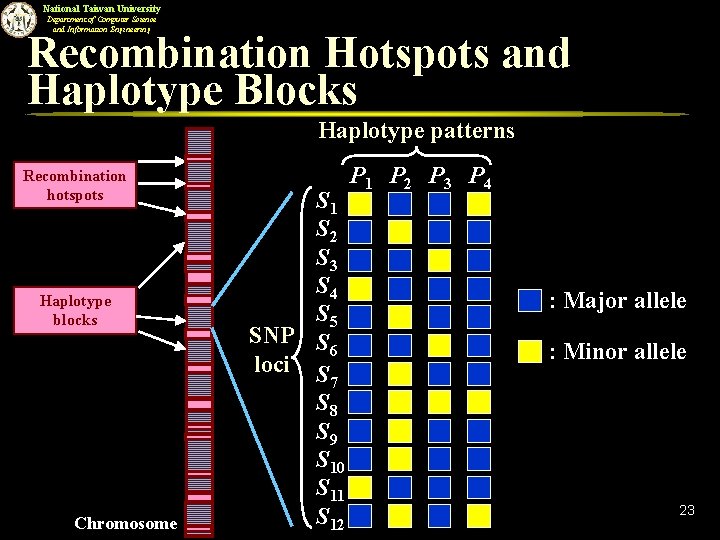
National Taiwan University Department of Computer Science and Information Engineering Haplotype Blocks and Tag SNPs n n Recent studies have shown that the chromosome can be partitioned into haplotype blocks interspersed by recombination hotspots (Daly et al, Patil et al. ). u Within a haplotype block, there is little or no recombination occurred. u The SNPs within a haplotype block tend to be inherited together. Within a haplotype block, a small subset of SNPs (called tag SNPs) is sufficient to distinguish each pair of haplotype patterns in the block. u We only need to genotype tag SNPs instead of all SNPs within a haplotype block. 22

National Taiwan University Department of Computer Science and Information Engineering Recombination Hotspots and Haplotype Blocks Haplotype patterns Recombination hotspots Haplotype blocks Chromosome S 1 S 2 S 3 S 4 S 5 SNP S 6 loci S 7 S 8 S 9 S 10 S 11 S 12 P 1 P 2 P 3 P 4 : Major allele : Minor allele 23

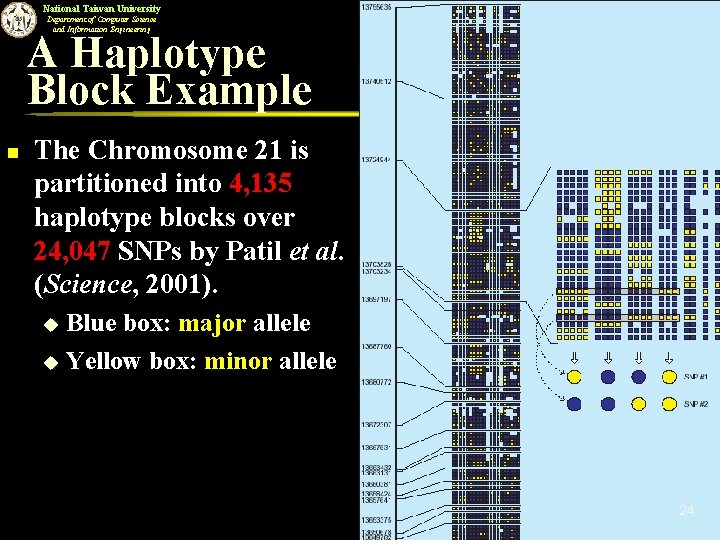
National Taiwan University Department of Computer Science and Information Engineering A Haplotype Block Example n The Chromosome 21 is partitioned into 4, 135 haplotype blocks over 24, 047 SNPs by Patil et al. (Science, 2001). Blue box: major allele u Yellow box: minor allele u 24

National Taiwan University Department of Computer Science and Information Engineering Examples of Tag SNPs Haplotype patterns S 1 S 2 S 3 S 4 S 5 SNP S 6 loci S 7 S 8 S 9 S 10 S 11 S 12 An unknown haplotype sample P 1 P 2 P 3 P 4 n n Suppose we wish to distinguish an unknown haplotype sample. We can genotype all SNPs to identify the haplotype sample. : Major allele : Minor allele 25

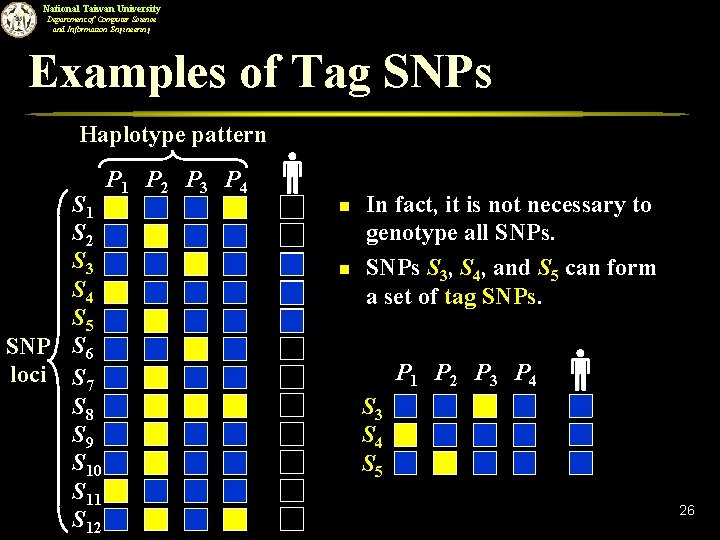
National Taiwan University Department of Computer Science and Information Engineering Examples of Tag SNPs Haplotype pattern S 1 S 2 S 3 S 4 S 5 SNP S 6 loci S 7 S 8 S 9 S 10 S 11 S 12 P 1 P 2 P 3 P 4 n n In fact, it is not necessary to genotype all SNPs S 3, S 4, and S 5 can form a set of tag SNPs. P 1 P 2 P 3 P 4 S 3 S 4 S 5 26

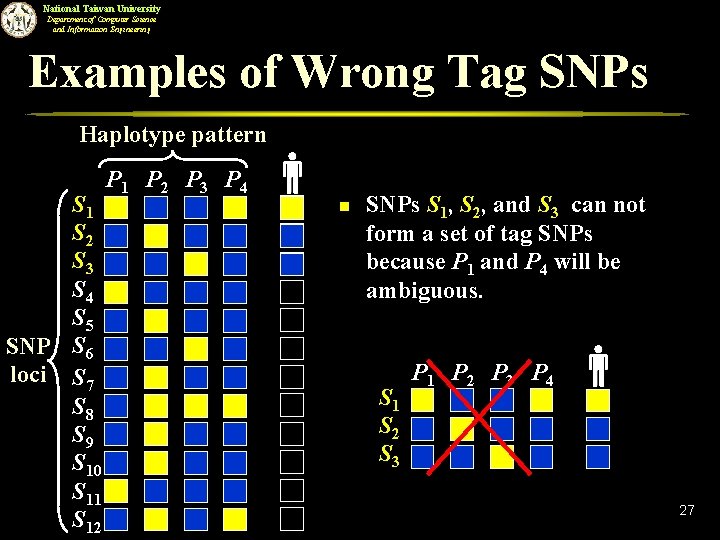
National Taiwan University Department of Computer Science and Information Engineering Examples of Wrong Tag SNPs Haplotype pattern S 1 S 2 S 3 S 4 S 5 SNP S 6 loci S 7 S 8 S 9 S 10 S 11 S 12 P 1 P 2 P 3 P 4 n SNPs S 1, S 2, and S 3 can not form a set of tag SNPs because P 1 and P 4 will be ambiguous. S 1 S 2 S 3 P 1 P 2 P 3 P 4 27

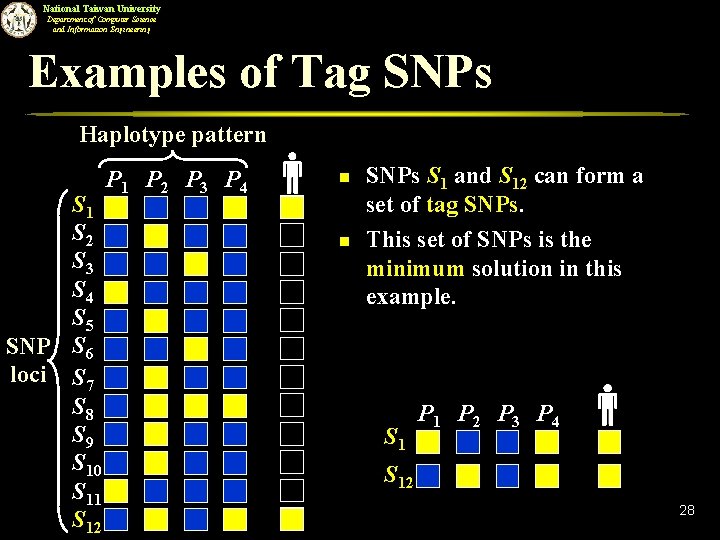
National Taiwan University Department of Computer Science and Information Engineering Examples of Tag SNPs Haplotype pattern S 1 S 2 S 3 S 4 S 5 SNP S 6 loci S 7 S 8 S 9 S 10 S 11 S 12 P 1 P 2 P 3 P 4 n n SNPs S 1 and S 12 can form a set of tag SNPs. This set of SNPs is the minimum solution in this example. S 1 P 2 P 3 P 4 S 12 28

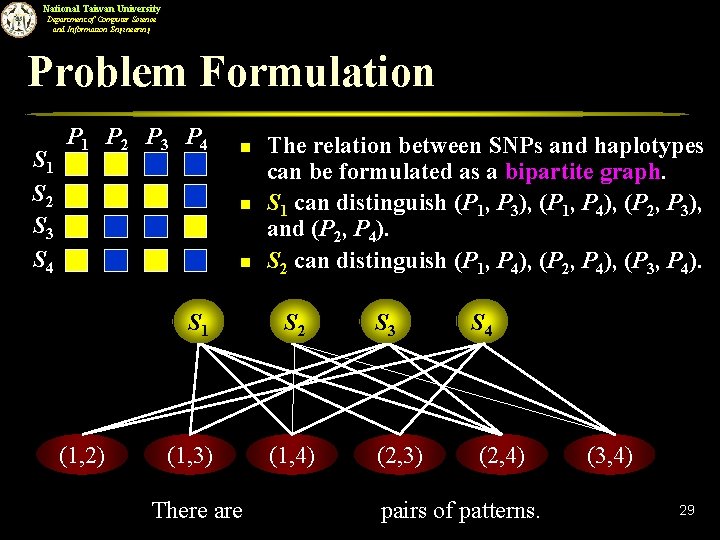
National Taiwan University Department of Computer Science and Information Engineering Problem Formulation S 1 S 2 S 3 S 4 P 1 P 2 P 3 P 4 n n n S 1 (1, 2) (1, 3) There are The relation between SNPs and haplotypes can be formulated as a bipartite graph. S 1 can distinguish (P 1, P 3), (P 1, P 4), (P 2, P 3), and (P 2, P 4). S 2 can distinguish (P 1, P 4), (P 2, P 4), (P 3, P 4). S 2 (1, 4) S 3 (2, 3) S 4 (2, 4) pairs of patterns. (3, 4) 29

National Taiwan University Department of Computer Science and Information Engineering Observation S 1 S 2 S 3 S 4 P 1 P 2 P 3 P 4 n n n S 1 (1, 2) (1, 3) The SNPs can form a set of tag SNPs if each pair of patterns is connected by at least one edge. e. g. , S 1 and S 3 can form a set of tag SNPs. e. g. , S 1 and S 2 can not be tag SNPs. S 2 (1, 4) S 3 (2, 3) (2, 4) (3, 4) Each pair of patterns is connected by at least one edge. 30

National Taiwan University Department of Computer Science and Information Engineering Problems of Finding Tag SNPs n n The problem of finding the minimum set of tag SNPs is known to be NP-hard. u This problem is the minimum test set problem. u A number of methods have been proposed to find the minimum set of tag SNPs (Bafna et al. , Zhang, et al. ). In reality, we may fail to obtain some tag SNPs if they do not pass the threshold of data quality. u In the current genotyping environment, the missing rate of SNPs is around 5~10%. u We proposed two greedy algorithms and one linear programming relaxation algorithm to solve this problem. 31

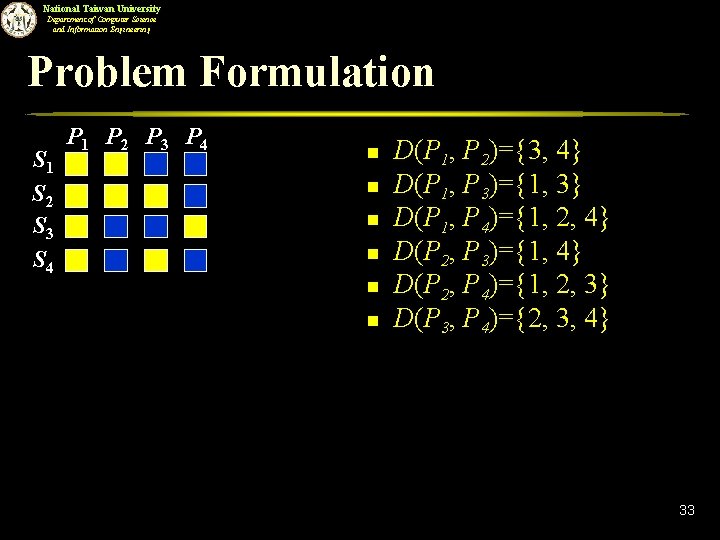
National Taiwan University Department of Computer Science and Information Engineering Integer Linear Programming n n N SNPs, K patterns Let xi be defined as follows. u u n n xi = 1 if the i-th SNP is selected; xi = 0 otherwise. Let D(Pi, Pj) be the set of SNPs that can distinguish patterns Pi and Pj. Integer programming formulation. 32

National Taiwan University Department of Computer Science and Information Engineering Problem Formulation S 1 S 2 S 3 S 4 P 1 P 2 P 3 P 4 n n n D(P 1, P 2)={3, 4} D(P 1, P 3)={1, 3} D(P 1, P 4)={1, 2, 4} D(P 2, P 3)={1, 4} D(P 2, P 4)={1, 2, 3} D(P 3, P 4)={2, 3, 4} 33

National Taiwan University Department of Computer Science and Information Engineering References: n n Huang, Y. -T. , Zhang, K. , Chen, T. and Chao, K. -M. , 2005, “Selecting Additional Tag SNPs for Tolerating Missing Data in Genotyping, ” BMC Bioinformatics, 6: 263. Chang, C. -J. , Huang, Y. -T. , and Chao, K. -M. , 2006, “A Greedier Approach for Finding Tag SNPs, ” Bioinformatics, 22: 685 -691. 34

National Taiwan University Department of Computer Science and Information Engineering Research Directions of SNPs and Haplotypes in Recent Years SNP Database Haplotype Inference Tag SNP Selection … Maximum Parsimony Perfect Phylogeny Statistical Methods Haplotype block LD bin Prediction Accuracy 35

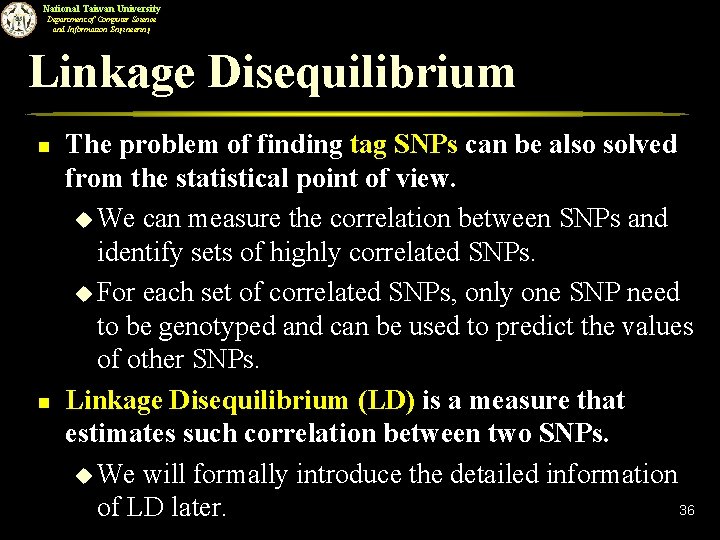
National Taiwan University Department of Computer Science and Information Engineering Linkage Disequilibrium n n The problem of finding tag SNPs can be also solved from the statistical point of view. u We can measure the correlation between SNPs and identify sets of highly correlated SNPs. u For each set of correlated SNPs, only one SNP need to be genotyped and can be used to predict the values of other SNPs. Linkage Disequilibrium (LD) is a measure that estimates such correlation between two SNPs. u We will formally introduce the detailed information 36 of LD later.

National Taiwan University Department of Computer Science and Information Engineering Linkage Disequilibrium Bins n The statistical methods for finding tag SNPs are based on the analysis of LD among all SNPs. An LD bin is a set of SNPs such that SNPs within the same bin are highly correlated with each other. u The value of a single SNP in one LD bin can predict the values of other SNPs of the same bin. u These methods try to identify the minimum set of LD bins. u 37

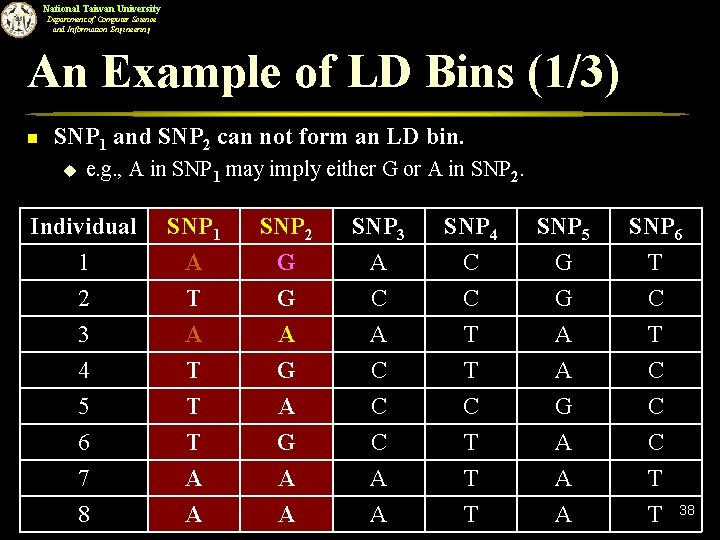
National Taiwan University Department of Computer Science and Information Engineering An Example of LD Bins (1/3) n SNP 1 and SNP 2 can not form an LD bin. u e. g. , A in SNP 1 may imply either G or A in SNP 2. Individual 1 2 3 SNP 1 A T A SNP 2 G G A SNP 3 A C A SNP 4 C C T SNP 5 G G A SNP 6 T C T 4 5 6 7 T T T A G A C C C A T C T T A G A A C C C T 8 A A A T 38

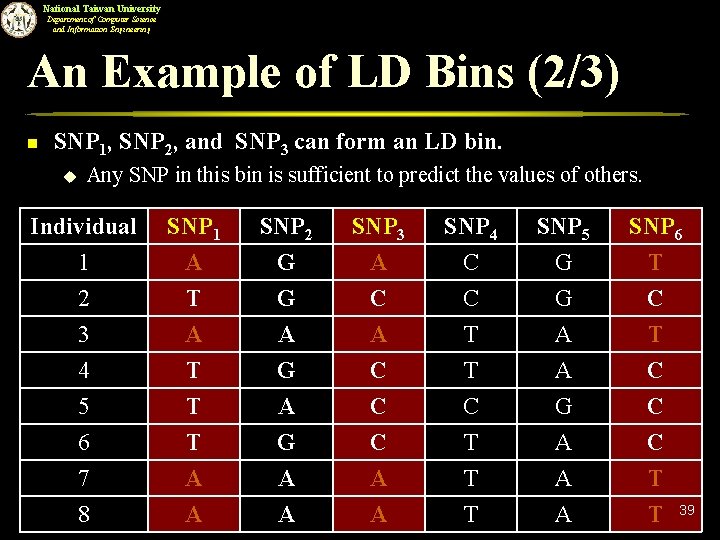
National Taiwan University Department of Computer Science and Information Engineering An Example of LD Bins (2/3) n SNP 1, SNP 2, and SNP 3 can form an LD bin. u Any SNP in this bin is sufficient to predict the values of others. Individual 1 2 3 SNP 1 A T A SNP 2 G G A SNP 3 A C A SNP 4 C C T SNP 5 G G A SNP 6 T C T 4 5 6 7 T T T A G A C C C A T C T T A G A A C C C T 8 A A A T 39

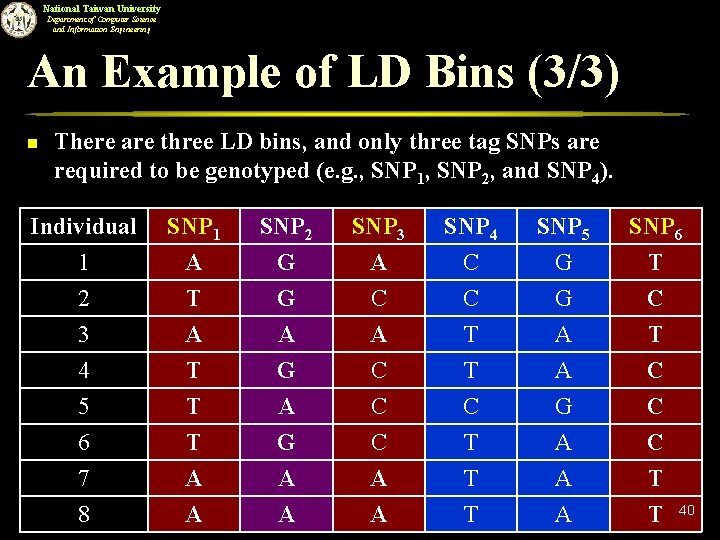
National Taiwan University Department of Computer Science and Information Engineering An Example of LD Bins (3/3) n There are three LD bins, and only three tag SNPs are required to be genotyped (e. g. , SNP 1, SNP 2, and SNP 4). Individual 1 2 3 SNP 1 A T A SNP 2 G G A SNP 3 A C A SNP 4 C C T SNP 5 G G A SNP 6 T C T 4 5 6 7 T T T A G A C C C A T C T T A G A A C C C T 8 A A A T 40

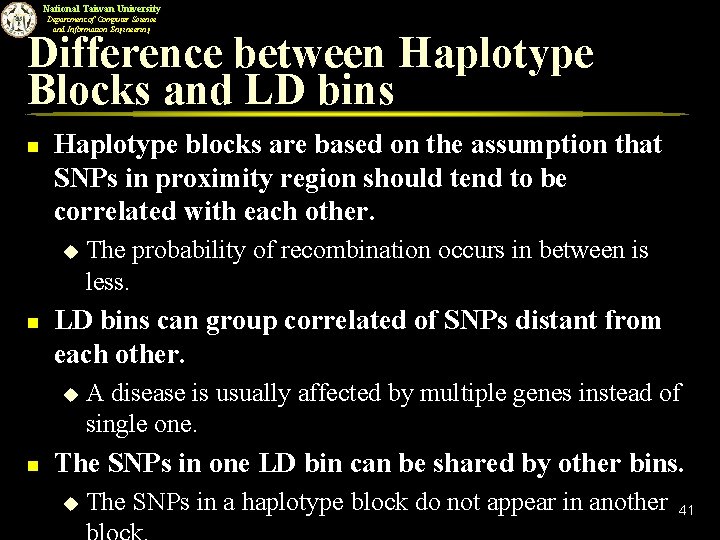
National Taiwan University Department of Computer Science and Information Engineering Difference between Haplotype Blocks and LD bins n Haplotype blocks are based on the assumption that SNPs in proximity region should tend to be correlated with each other. u n LD bins can group correlated of SNPs distant from each other. u n The probability of recombination occurs in between is less. A disease is usually affected by multiple genes instead of single one. The SNPs in one LD bin can be shared by other bins. u The SNPs in a haplotype block do not appear in another 41

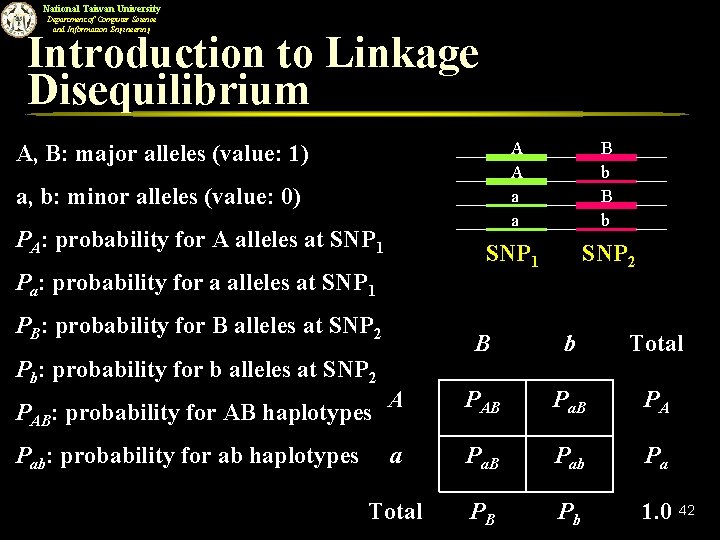
National Taiwan University Department of Computer Science and Information Engineering Introduction to Linkage Disequilibrium A A a a B b SNP 1 SNP 2 A, B: major alleles (value: 1) a, b: minor alleles (value: 0) PA: probability for A alleles at SNP 1 Pa: probability for a alleles at SNP 1 PB: probability for B alleles at SNP 2 B b Total A PAB Pa. B PA a Pa. B Pab Pa Total PB Pb 1. 0 Pb: probability for b alleles at SNP 2 PAB: probability for AB haplotypes Pab: probability for ab haplotypes 42

National Taiwan University Department of Computer Science and Information Engineering Linkage Equilibrium n n PAB = PAPB PAb = PAPb = PA(1 -PB) Pa. B = Pa. PB = (1 -PA) PB Pab = Pa. Pb = (1 -PA) (1 -PB) SNP 1 SNP 2 B b Total A PAB Pa. B PA a Pa. B Pab Pa Total PB Pb 1. 0 43

National Taiwan University Department of Computer Science and Information Engineering Linkage Disequilibrium n n PAB ≠ PAPB PAb ≠ PAPb = PA(1 -PB) Pa. B ≠ Pa. PB = (1 -PA) PB Pab ≠ Pa. Pb = (1 -PA) (1 -PB) SNP 1 SNP 2 B b Total A PAB Pa. B PA a Pa. B Pab Pa Total PB Pb 1. 0 44

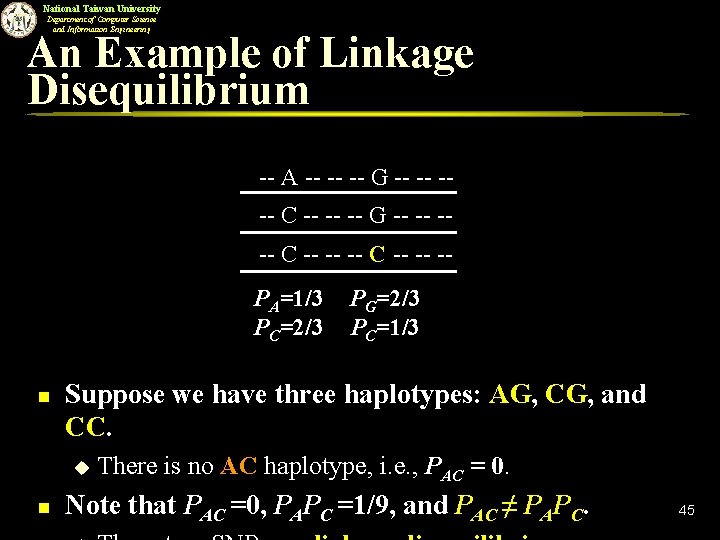
National Taiwan University Department of Computer Science and Information Engineering An Example of Linkage Disequilibrium -- A -- -- -- G -- -- --- C -- -- -PA=1/3 PC=2/3 n Suppose we have three haplotypes: AG, CG, and CC. u n PG=2/3 PC=1/3 There is no AC haplotype, i. e. , PAC = 0. Note that PAC =0, PAPC =1/9, and PAC ≠ PAPC. 45

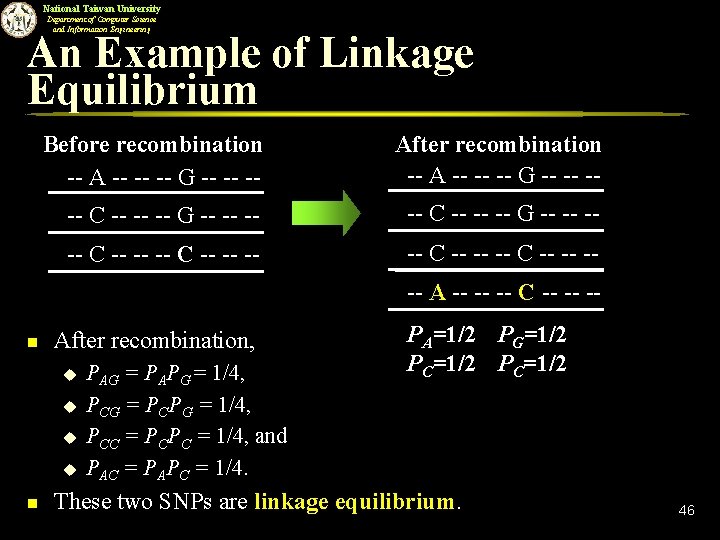
National Taiwan University Department of Computer Science and Information Engineering An Example of Linkage Equilibrium Before recombination -- A -- -- -- G -- -- -- After recombination -- A -- -- -- G -- -- -- -- C -- -- -- C -- -- --- A -- -- -- C -- -- -- n After recombination, u u n PAG = PAPG = 1/4, PCG = PCPG = 1/4, PCC = PCPC = 1/4, and PAC = PAPC = 1/4. PA=1/2 PG=1/2 PC=1/2 These two SNPs are linkage equilibrium. 46

National Taiwan University Department of Computer Science and Information Engineering Linkage Disequilibrium n There are many formulas to compute LD between two SNPs, and most of them are usually normalized between -1~1 or 0~1. u LD = 1 (perfect positive correlation) u LD = 0 (no correlation or linkage equilibrium) u LD = -1 (perfect negative correlation) u LD = 0. 8 (strong positive correlation) u LD = 0. 12 (weak positive correlation) 47

National Taiwan University Department of Computer Science and Information Engineering Linkage Disequilibrium Formulas n Mathematical formulas for computing LD: u r 2 u or Δ 2: D’: Chi-square Test. u P value. u 48

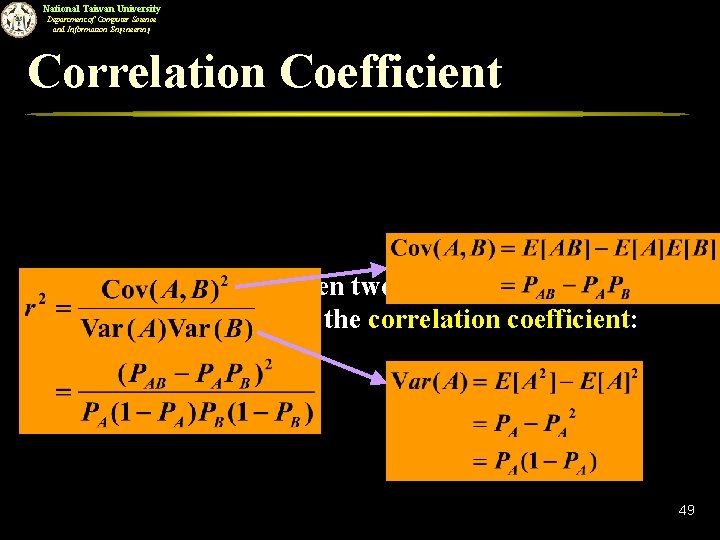
National Taiwan University Department of Computer Science and Information Engineering Correlation Coefficient n The correlation between two random variables A and B can be measured by the correlation coefficient: 49

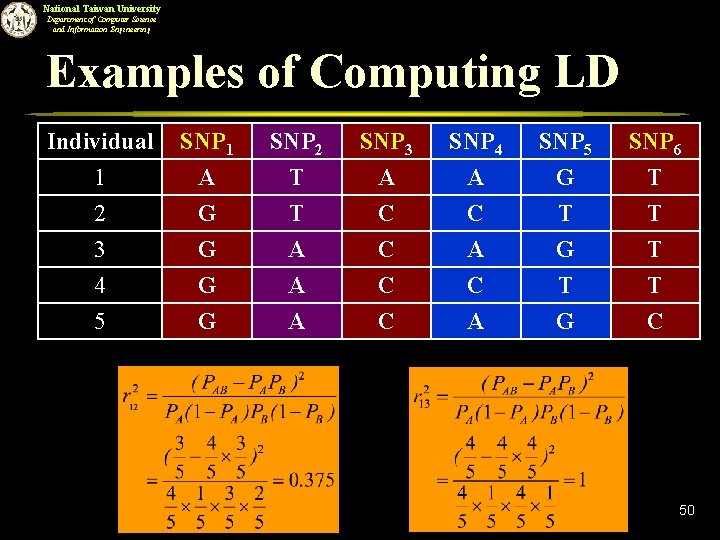
National Taiwan University Department of Computer Science and Information Engineering Examples of Computing LD Individual 1 2 3 SNP 1 A G G SNP 2 T T A SNP 3 A C C SNP 4 A C A SNP 5 G T G SNP 6 T T T 4 5 G G A A C C C A T G T C 50

National Taiwan University Department of Computer Science and Information Engineering Minimum Clique Cover Problem n This problem asks for a minimum set of LD bins. u n The minimum LD value required between two SNPs in one bin is usually set to 0. 8. This problem is known to be the minimum clique cover problem (by Huang and Chao, 2005). Consider each SNP as nodes on the graph. u There exists an edge between two nodes iff the LD of these two SNPs ≥ 0. 8. u 51

National Taiwan University Department of Computer Science and Information Engineering Relaxation of This Problem n The minimum clique cover problem is not easy to be approximated. u n The relaxed problem asks for a minimum set of LD bins such that at least one SNP in an LD bin has r 2 ≥ 0. 8 with other SNPs in the same bin. The relaxed problem is known to be the minimum dominating set problem. u The minimum dominating set problem is still NP-hard but is easier to be approximated. 52

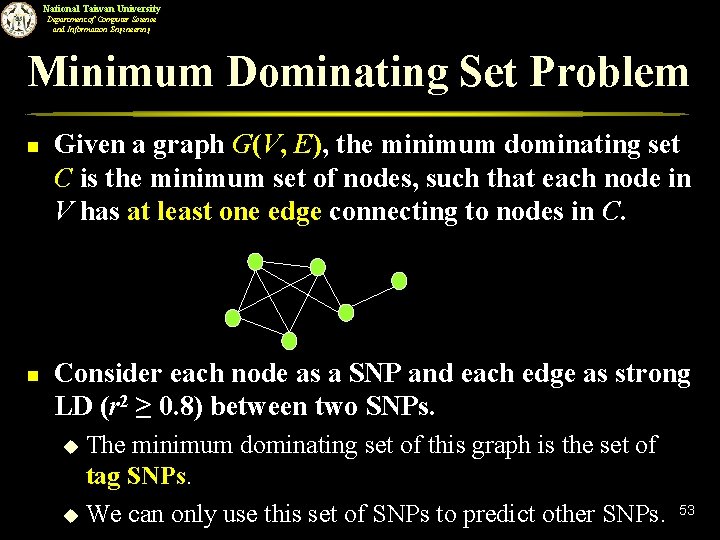
National Taiwan University Department of Computer Science and Information Engineering Minimum Dominating Set Problem n n Given a graph G(V, E), the minimum dominating set C is the minimum set of nodes, such that each node in V has at least one edge connecting to nodes in C. Consider each node as a SNP and each edge as strong LD (r 2 ≥ 0. 8) between two SNPs. The minimum dominating set of this graph is the set of tag SNPs. u We can only use this set of SNPs to predict other SNPs. u 53

National Taiwan University Department of Computer Science and Information Engineering Experimental Data Sets n Hinds et al. (2005) identified 1, 586, 383 SNPs across three human populations. u n African, Americans of European, and Asian. The database provides both genotype data and inferred haplotype data. 54
 Ntu ce
Ntu ce National science council taiwan
National science council taiwan Columbia university department of computer science
Columbia university department of computer science Seoul national university computer science
Seoul national university computer science National yunlin university of science and technology
National yunlin university of science and technology Providence university taiwan ranking
Providence university taiwan ranking Ucl university computer science
Ucl university computer science Northwestern computer science department
Northwestern computer science department Computer science department rutgers
Computer science department rutgers Meredith hutchin
Meredith hutchin Fsu cybersecurity major
Fsu cybersecurity major Trimentoring
Trimentoring Bhargavi goswami
Bhargavi goswami Science my favourite subject
Science my favourite subject Phoenix online computer science university
Phoenix online computer science university University of bridgeport engineering
University of bridgeport engineering University of bridgeport computer science
University of bridgeport computer science Yonsei computer science
Yonsei computer science York university computer science
York university computer science Unc ch computer science
Unc ch computer science Osaka university computer science
Osaka university computer science Computer science columbia university
Computer science columbia university 7800 york road
7800 york road Kansas state university computer science
Kansas state university computer science Brown university computer science faculty
Brown university computer science faculty Trinity university computer science
Trinity university computer science Brandeis university computer science
Brandeis university computer science Institute of computer science university of tartu
Institute of computer science university of tartu Kotebe metropolitan university website
Kotebe metropolitan university website Colonialism and development: korea, taiwan, and kwantung
Colonialism and development: korea, taiwan, and kwantung National risk and resilience ambulance
National risk and resilience ambulance Eacads iitd
Eacads iitd Tum
Tum National core standards priorities
National core standards priorities National audit department
National audit department National unification and the national state
National unification and the national state Department of law university of jammu
Department of law university of jammu Department of geology university of dhaka
Department of geology university of dhaka Narrativistic
Narrativistic University of bridgeport it department
University of bridgeport it department University of iowa math
University of iowa math Sputonik v
Sputonik v Texas state university psychology
Texas state university psychology Department of information engineering university of padova
Department of information engineering university of padova Department of information engineering university of padova
Department of information engineering university of padova Manipal university chemistry department
Manipal university chemistry department Syracuse university psychology department
Syracuse university psychology department Jackson state university finance department
Jackson state university finance department Michigan state astronomy
Michigan state astronomy Columbia university cs department
Columbia university cs department University of sargodha engineering department
University of sargodha engineering department Claudia arrighi
Claudia arrighi Kyiv national university of culture and arts
Kyiv national university of culture and arts National and kapodistrian university of athens events
National and kapodistrian university of athens events Department of forensic science dc
Department of forensic science dc