n Sequence homology similarity and comparison Homology is























































- Slides: 87


n Sequence homology, similarity and comparison 序列同源性、相似性和序列比对 'Homology' is one of the most important terms in biology. 生物信息学 2


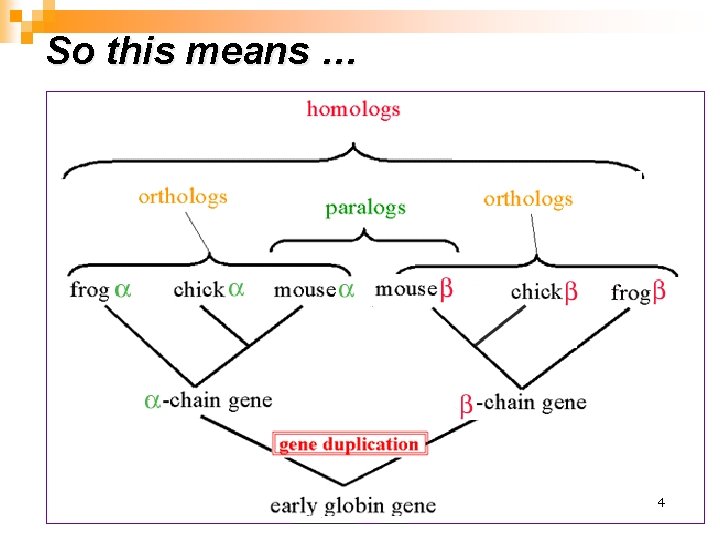
So this means … 4 生物信息学 4

利用orthologous构建不同物种的系统发育树 “Phylogenetic(系统发育的) reconstructions of organisms created using information from the nucleotide sequences of genes require orthologous, rather than paralogous genes, so the distinction between these two gene classes is important for practical reasons. ” 生物信息学 5


Homology 同源性 n n n Features derived from a common ancestor are called homologs. New sequences are adapted from pre-existing sequences rather than invented de novo (从新 开始). Nature is a tinkerer and not an inventor. Its products are not necessarily neat or elegant. (Jacob. 1977. Science 196: 1161 -1166) 进化是一位修补匠, 而不是发明家。他的产物不必 整洁而又优雅. 生物信息学 7

Assumption: genetic constitution of organisms can be traced back to a set of common ancestral genes. 假设:通过追溯一系列共同祖先基因,我们可以构建 物种之间的亲缘关系。 n Thus, we can make a comparison between gene sequences from different species to identify the distances between them. 基于上面的假设,我们可以通过比较不同物种的同 源序列的差异,来推断这些物种或者序列之间的进 化距离。 生物信息学 8

Homology Similarity Orthologous relationships: p One to one ? One to many? Or Many to many? p Complex: gene duplication, gene loss and speciation can be frequent events in the history of a group of organisms. 基因复制、基因丢失和物种分化等进化事件频繁发生,导 致不同物种的同源基因数量很不一致。 Genetic homology is inferred from significant similarity; Similarity however does not necessarily imply homology. 生物信息学 9

Further reading n Fitch WM. (2000) Homology - a personal view on some of the problems. TRENDS IN GENETICS 16 (5): 227 -231. n Sonnhammer ELL and Koonin EV. (2002) Orthology, paralogy and proposed classification for paralog subtypes. TRENDS IN GENETICS 18 (12): 619 -620. 这两篇文献不提供PDFs,你们利用Pub. Med或者其他搜索引擎来 搜索文献。依据个人习惯,自由选择在线阅读,或者下载PDF 阅读。 生物信息学 10



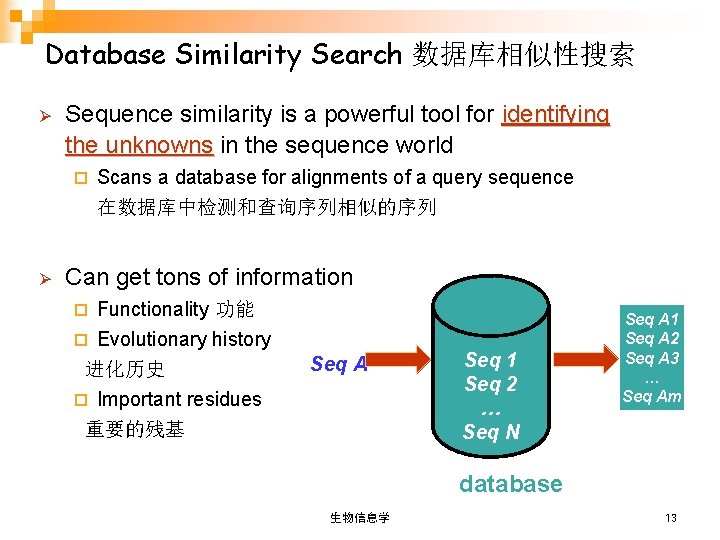
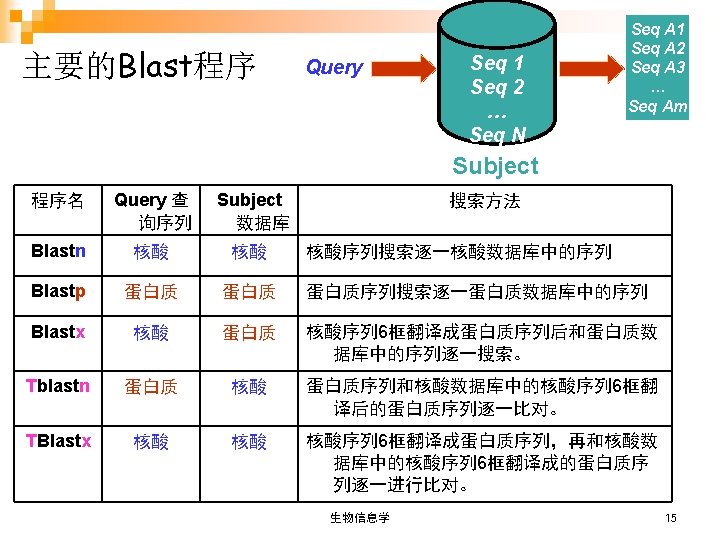
Database Similarity Search 数据库相似性搜索 Ø Sequence similarity is a powerful tool for identifying the unknowns in the sequence world ¨ Ø Scans a database for alignments of a query sequence 在数据库中检测和查询序列相似的序列 Can get tons of information Functionality 功能 ¨ Evolutionary history 进化历史 ¨ Important residues 重要的残基 ¨ Seq A Seq 1 Seq 2 … Seq N Seq A 1 Seq A 2 Seq A 3 … Seq Am database 生物信息学 13

Blast n n Blast 是“基本的局部相似性查询 具”(Basic Local Alignment Search Tool)的 缩写. ¨ Altschul SF, Gish W, Miller W, Myers EW & Lipman DJ. 1990. Basic local alignment search tool. JMB 215: 403 -415 ¨ Altschul & Gish 1996. Methods in Enzymology 266: 460 -480; ¨ Altschul et al. 1997. NAR 25: 3389 -3402 Blast 是一个序列相似性搜索的程序包,其中包含了很多个独立的程 序,这些程序是根据查询的对象和数据库的不同来定义的。比如说查 询的序列为核酸,查询数据库亦为核酸序列数据库,那么就应该选择 blastn程序。 n Fast & Heuristic (运行速度快&直观的) ¨ Not 100% assurance, but excellent in most cases. 生物信息学 14



Blast资源 1. NCBI主站点: http: //blast. ncbi. nlm. nih. gov (网络版) ftp: //ftp. ncbi. nlm. nih. gov/blast/ (单机版;本课程不讲授) 其他站点 http: //www. arabidopsis. org/Blast/index. jsp (拟南芥) http: //flybase. org/blast/ (果蝇) …… 2. 生物信息学 17

例子:Human Hemoglobin subunit beta ( 血红蛋白β亚基) n 对应的蛋白质序列: n >sp|P 68871|HBB_HUMAN Hemoglobin subunit beta OS=Homo sapiens GN=HBB PE=1 SV=2 MVHLTPEEKSAVTALWGKVNVDEVGGEALGRLLVVYPWTQRF FESFGDLSTPDAVMGNPK VKAHGKKVLGAFSDGLAHLDNLKGTFATLSELHCDKLHVDPEN FRLLGNVLVCVLAHHFG KEFTPPVQAAYQKVVAGVANALAHKYH 生物信息学 18

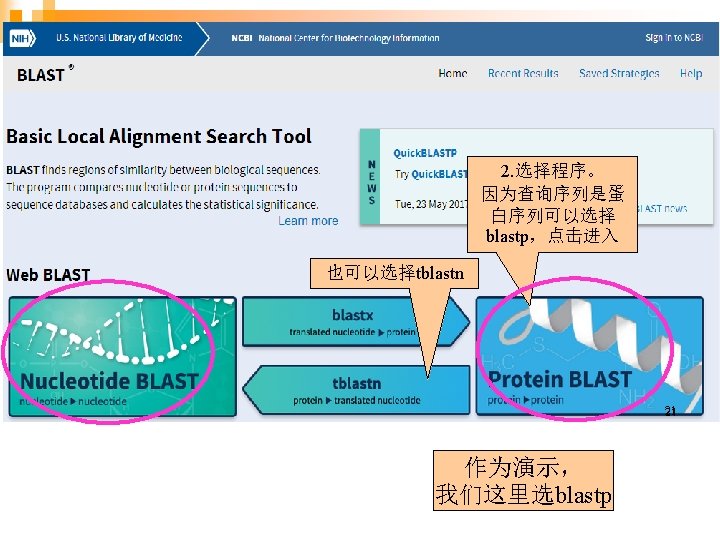
两两序列比对 (Pairwise alignment) http: //blast. ncbi. nlm. nih. gov 19



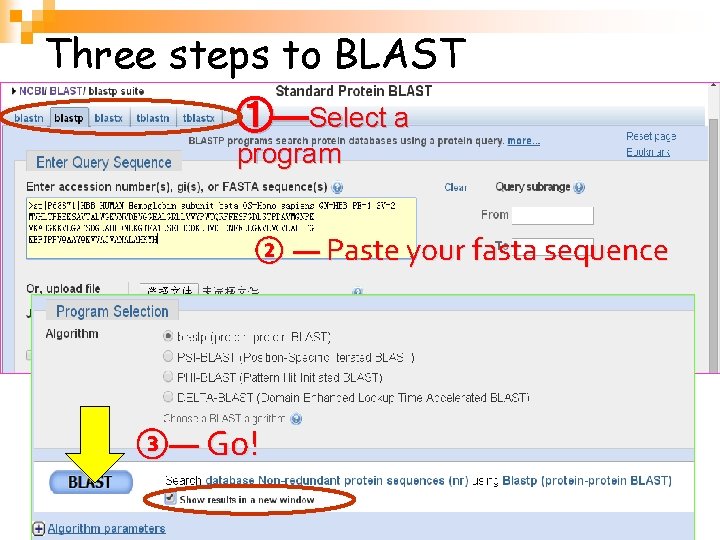
Three steps to BLAST ①—Select a program ② — Paste your fasta sequence ③— Go! 生物信息学 22

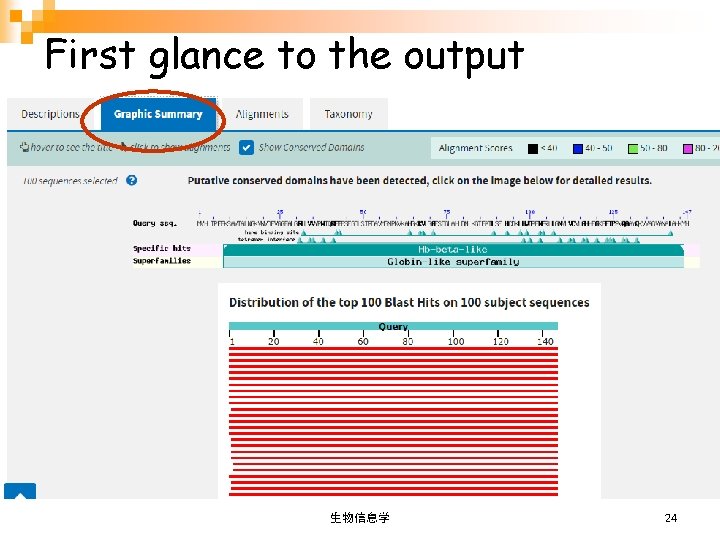
First glance to the output Job summary Four sections 生物信息学 23

First glance to the output 生物信息学 24

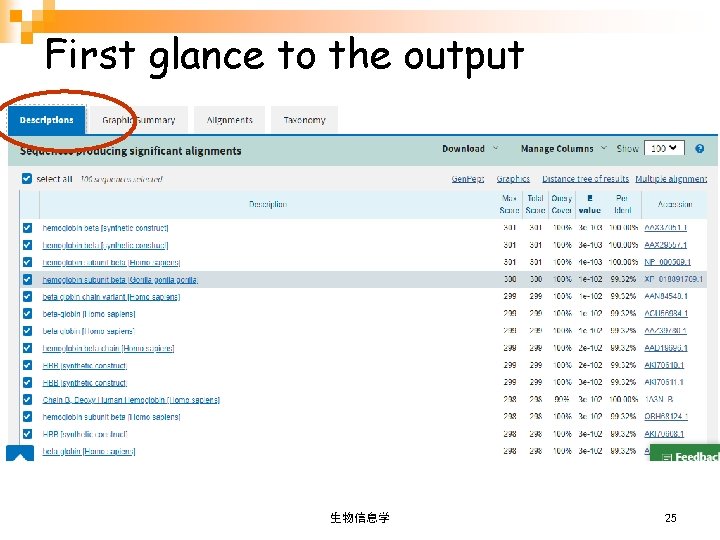
First glance to the output 生物信息学 25

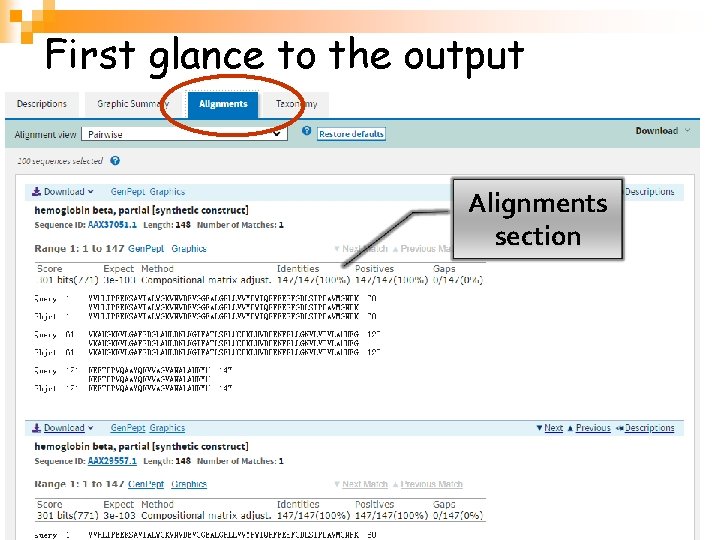
First glance to the output Alignments section

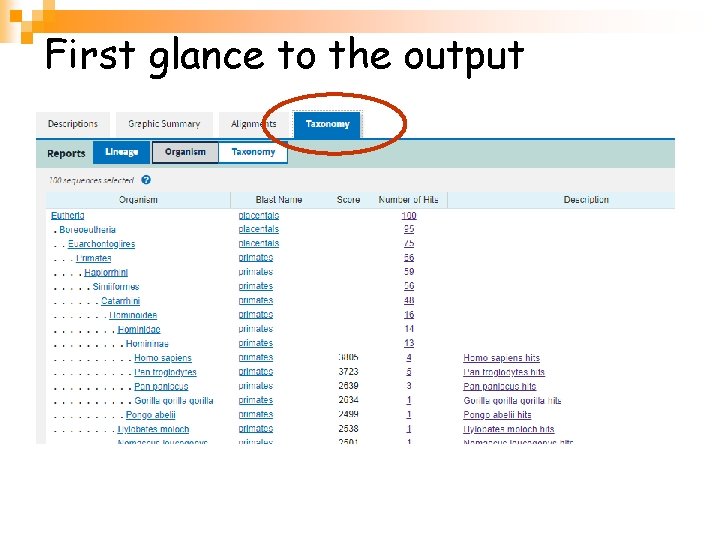
First glance to the output

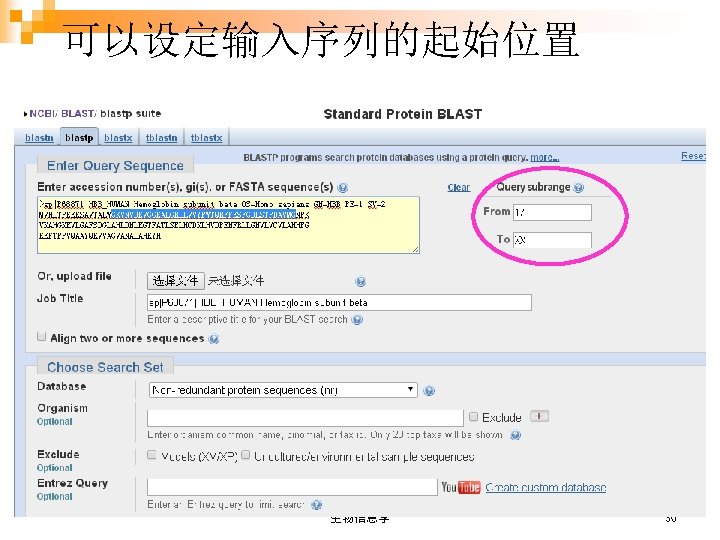
And now, more details… What if my sequence is saved in a fasta file, or my friend just tell me an accession number? ……………… 生物信息学 7 xx 28



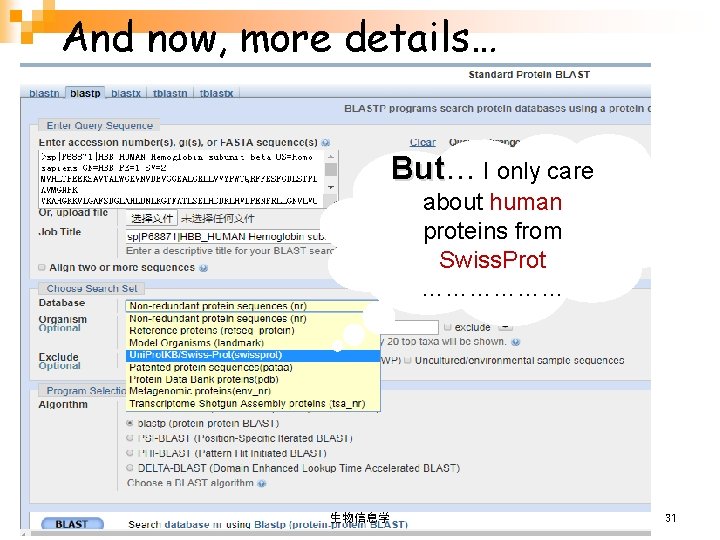
And now, more details… But I only care about human proteins from Swiss. Prot ……………… 生物信息学 31

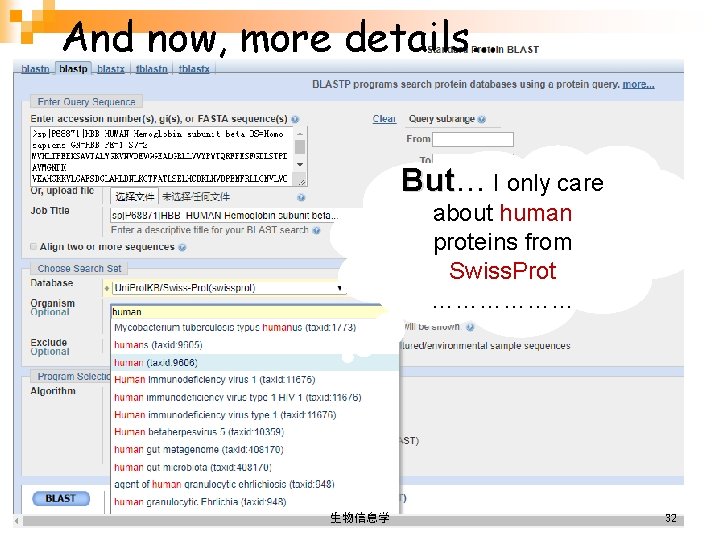
And now, more details… But I only care about human proteins from Swiss. Prot ……………… 生物信息学 32

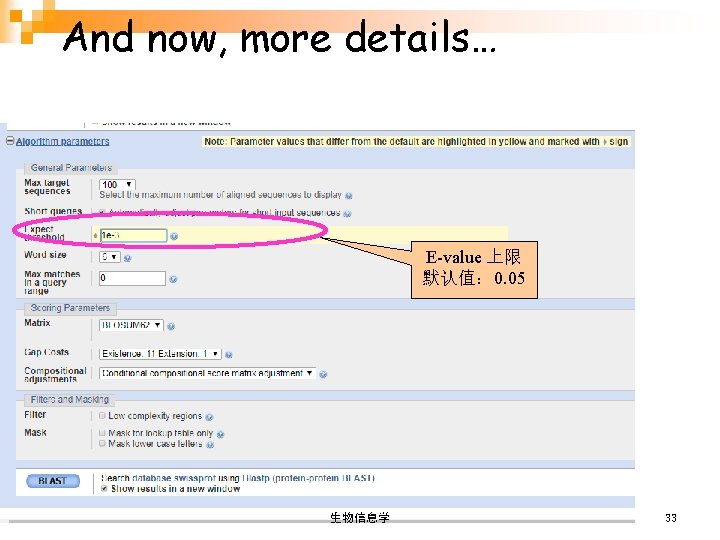
And now, more details… E-value 上限 默认值: 0. 05 生物信息学 33


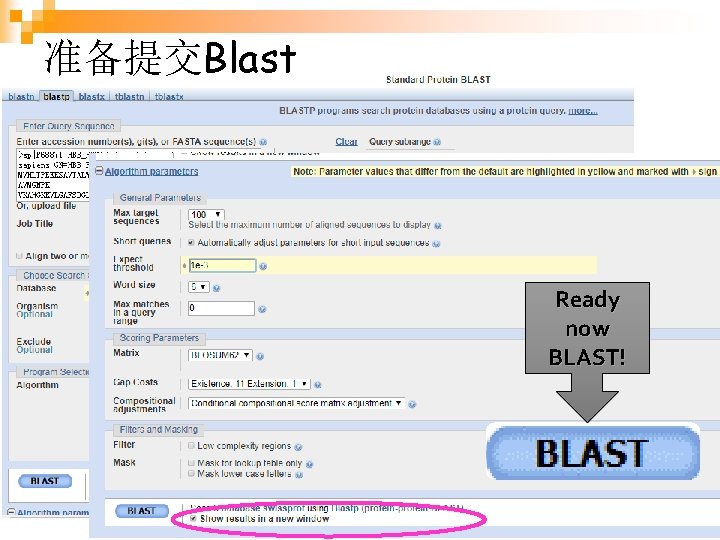
准备提交Blast Ready now BLAST! 生物信息学 35

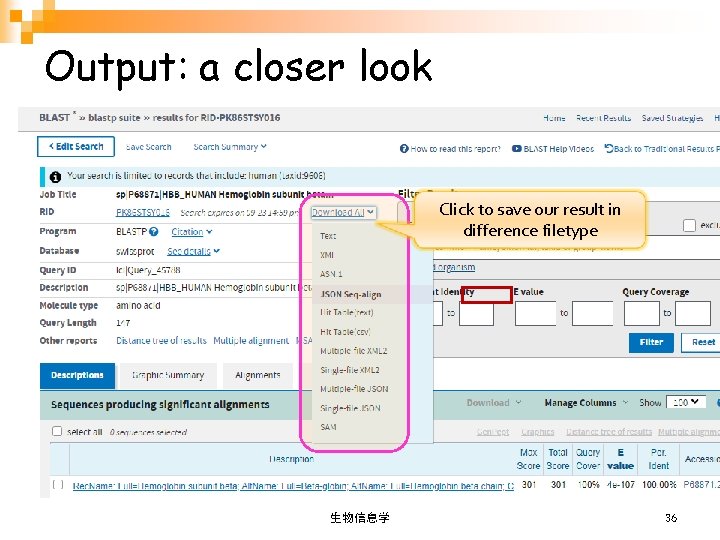
Output: a closer look Click to save our result in difference filetype 生物信息学 36


One-line description Human protein sequence from Swissprot(sp), with linkage to the protein webpage E-Value 表示因随 机性获得这一比对 结果的可能性 (值 越小越好) Bits score of each alignment(值越大越好 ) 生物信息学 38

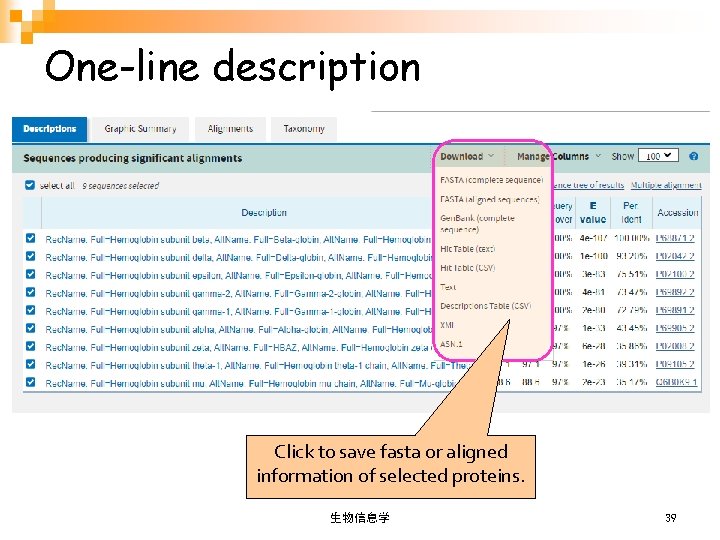
One-line description Click to save fasta or aligned information of selected proteins. 生物信息学 39

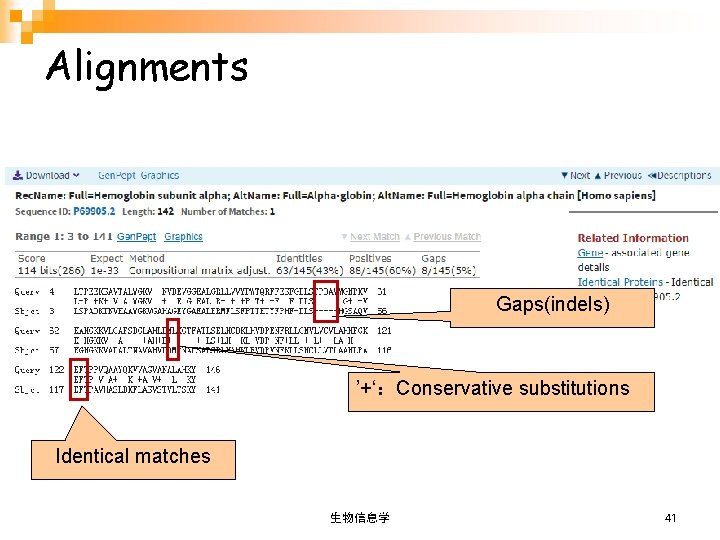
Alignments Sequence definition Sequence identifier l. Identities 序列相似性: Number of identical residues / length of alignment; l. Positives 序列一致性: Number of conservative substitutions / length of alignment; l. Gaps: Number of gaps / length of alignment. 生物信息学 40

Alignments Gaps(indels) ’+‘:Conservative substitutions Identical matches 生物信息学 41




Blast Help 生物信息学 45

作业 1. 重点:熟悉Blast运行的例子,熟悉Blast使 用流程和结果分析。 2. 可选:通过Blast Help了解更多内容。 http: //blast. ncbi. nlm. nih. gov/Blast. cgi? CMD=Web&PAGE_TYPE=Blast. Docs 生物信息学 46







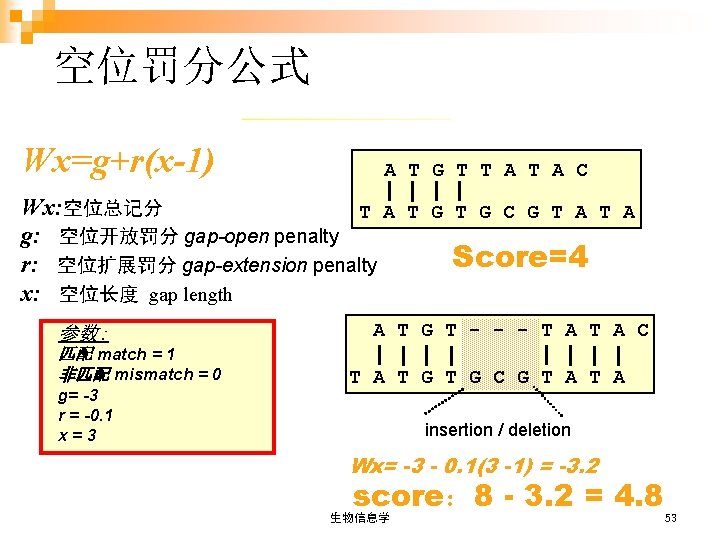
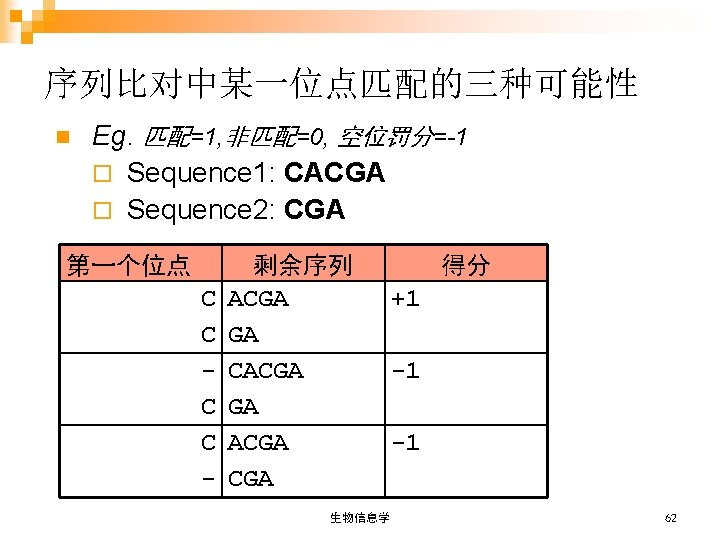
空位罚分公式 Wx=g+r(x-1) A T G T T A C Wx: 空位总记分 T A T G C G T A g: 空位开放罚分 gap-open penalty Score=4 r: 空位扩展罚分 gap-extension penalty x: 空位长度 gap length 参数: 匹配 match = 1 非匹配 mismatch = 0 g= -3 r = -0. 1 x=3 A T G T - - - T A C T A T G C G T A insertion / deletion Wx= -3 - 0. 1(3 -1) = -3. 2 score: 8 - 3. 2 = 4. 8 生物信息学 53

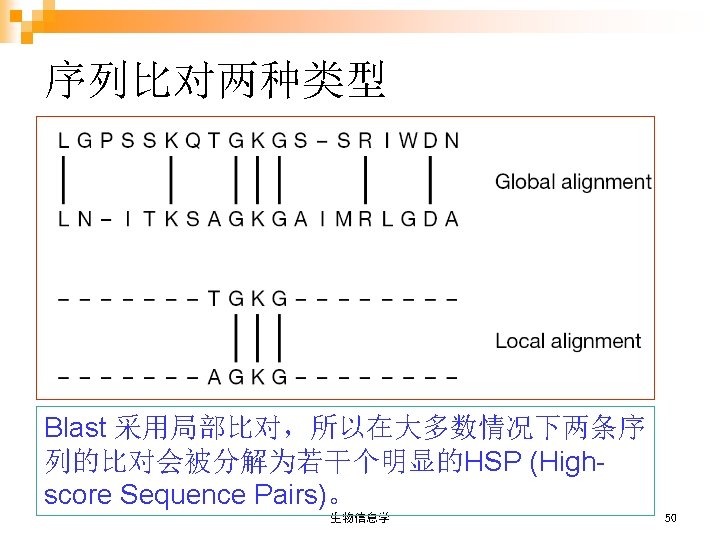
双序列比对方法 n 点阵序列比较 (Dot Matrix Sequence Comparison) n 动态规划算法 (Dynamic Programming Algorithm) n 词或K串方法 (Word or K-tuple Methods):不讲授 生物信息学 54


点阵法:自身的比对 A K G A 1 0 0 K 1 0 G 1 F K C A D E F 0 0 0 1 K 0 1 0 0 1 生物信息学 C 0 0 0 1 A 1 0 0 0 1 D 0 0 0 0 1 E 0 0 0 0 1 56

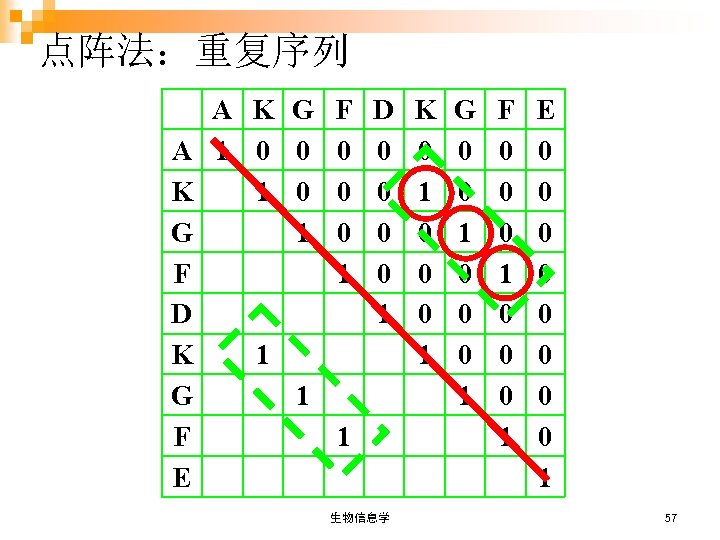
点阵法:重复序列 A K G A 1 0 0 K 1 0 G 1 F D K 1 G 1 F E F 0 0 0 1 D 0 0 1 1 生物信息学 K 0 1 0 0 0 1 G 0 0 1 0 0 0 1 F 0 0 0 1 E 0 0 0 0 1 57

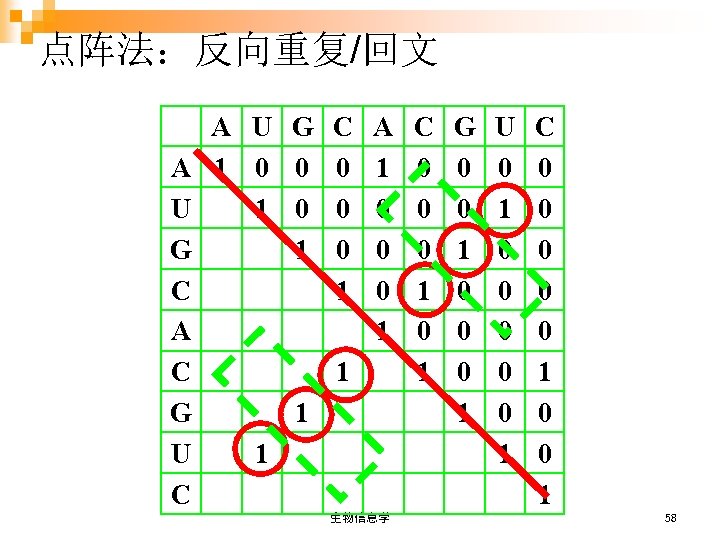
点阵法:反向重复/回文 A U G A 1 0 0 U 1 0 G 1 C A C G 1 U 1 C C 0 0 0 1 A 1 0 0 0 1 1 生物信息学 C 0 0 0 1 G 0 0 1 0 0 0 1 U 0 1 0 0 0 1 C 0 0 0 1 58

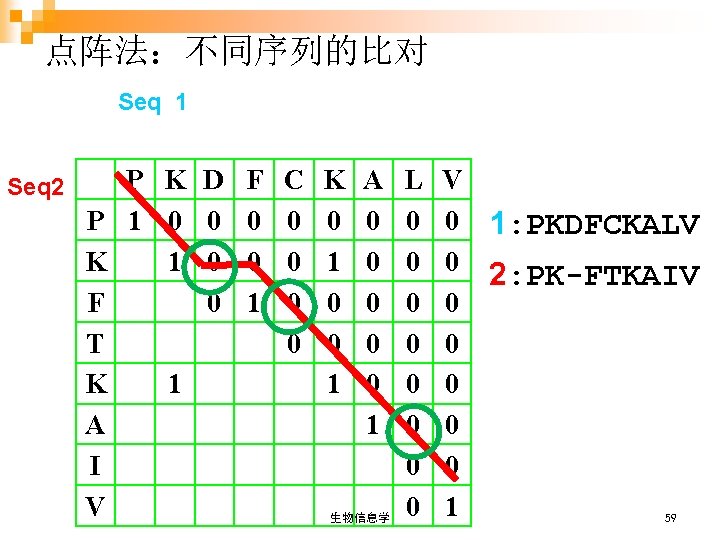
点阵法:不同序列的比对 Seq 1 Seq 2 P K D P 1 0 0 K 1 0 F 0 T K 1 A I V F 0 0 1 C 0 0 K 0 1 0 0 1 A 0 0 0 1 生物信息学 L 0 0 0 0 V 0 1: PKDFCKALV 0 2: PK-FTKAIV 0 0 0 1 59

点阵法的序列比对 Sequence 1# Sequence 2# 1 n 1 “-” Insertion m 生物信息学 60














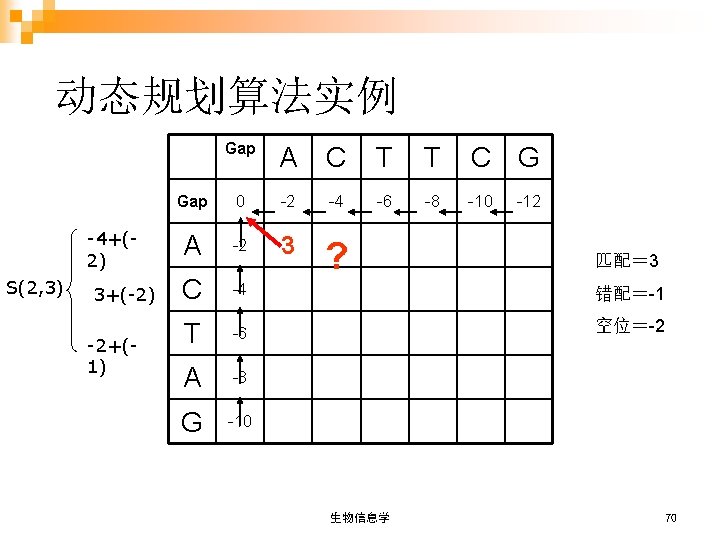
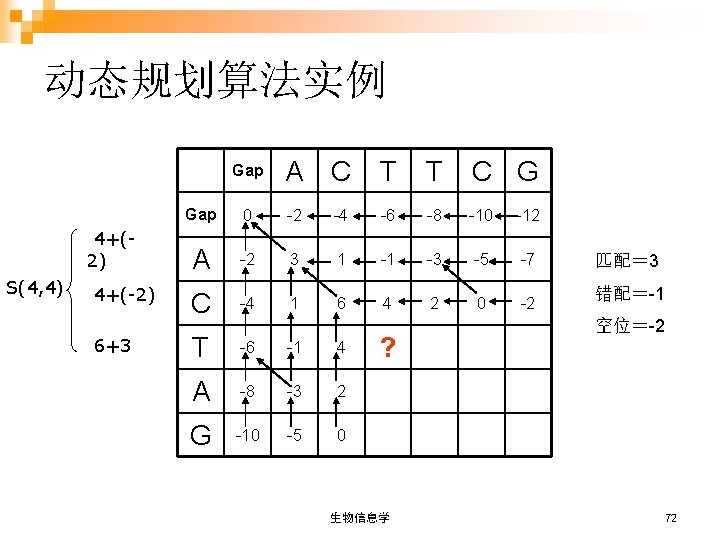
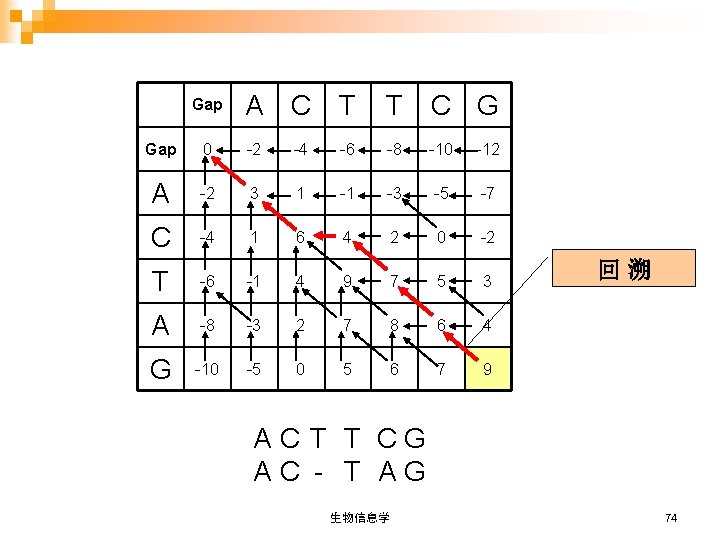
Gap A C T T C G Gap 0 -2 -4 -6 -8 -10 -12 A -2 3 1 -1 -3 -5 -7 C -4 1 6 4 2 0 -2 T -6 -1 4 9 7 5 3 A -8 -3 2 7 8 6 4 G -10 -5 0 5 6 7 9 回溯 AC T T CG AC - T AG 生物信息学 74

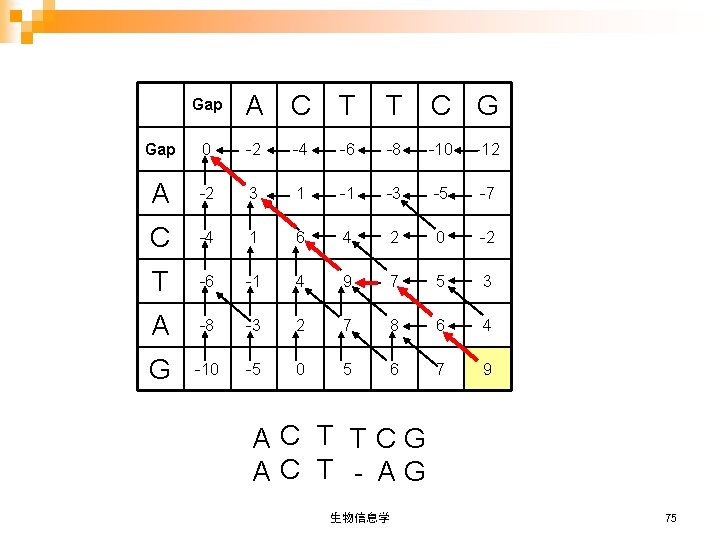
Gap A C T T C G Gap 0 -2 -4 -6 -8 -10 -12 A -2 3 1 -1 -3 -5 -7 C -4 1 6 4 2 0 -2 T -6 -1 4 9 7 5 3 A -8 -3 2 7 8 6 4 G -10 -5 0 5 6 7 9 AC T TCG AC T - AG 生物信息学 75

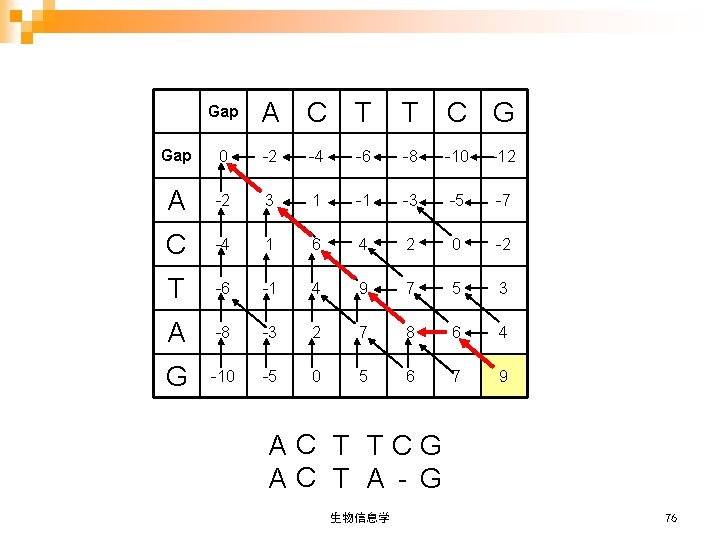
Gap A C T T C G Gap 0 -2 -4 -6 -8 -10 -12 A -2 3 1 -1 -3 -5 -7 C -4 1 6 4 2 0 -2 T -6 -1 4 9 7 5 3 A -8 -3 2 7 8 6 4 G -10 -5 0 5 6 7 9 AC T TCG AC T A - G 生物信息学 76

比对结果 1. ACTTCG AC-TAG 2. ACTTCG ACT-AG 3. ACTTCG ACTA-G 哪一个是最优比对 (optimal alignment)呢? 记分矩阵 生物信息学 77

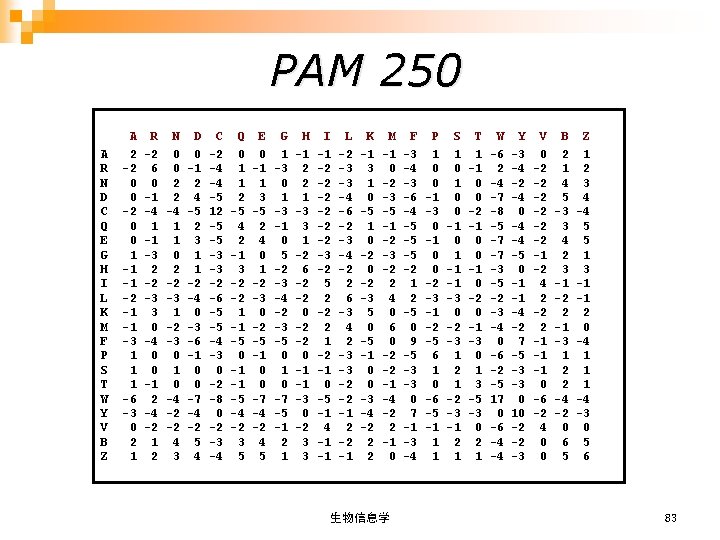
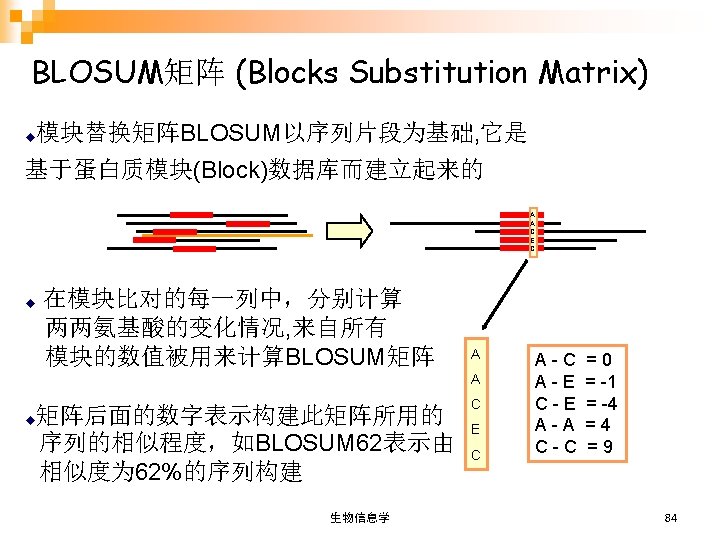
记分矩阵 (SCORING MATRICES) n DNA Scoring Matrices (DNA积分矩阵) n Amino Acid Substitution Matrices (氨基酸替换矩阵) PAM (Point Accepted Mutation) ¨ BLOSUM (Blocks Substitution Matrix) ¨ 生物信息学 78



蛋白质计分矩阵 Sequence 1 PTHPLASKTQILPEDLASEDLTI Sequence 2 PTHPLAGERAIGLARLAEEDFGM 记分矩阵 C S T P G N D . AT: T = 5 = -2. T: G Score = 48 C 9 S -1 4 T -1 1 5 P -3 -1 -1 7 A 0 1 0 -1 4 G -3 0 -2 -2 0 1 生物信息学 0 -2 -2 6 N -3 81


PAM 250 A R N D C Q E G H I L K M F P S T W Y V B Z A 2 -2 0 0 1 -1 -1 -2 -1 -1 -3 1 1 1 -6 -3 0 2 1 R -2 6 0 -1 -4 1 -1 -3 2 -2 -3 3 0 -4 0 0 -1 2 -4 -2 1 2 N 0 0 2 2 -4 1 1 0 2 -2 -3 1 -2 -3 0 1 0 -4 -2 -2 4 3 D 0 -1 2 4 -5 2 3 1 1 -2 -4 0 -3 -6 -1 0 0 -7 -4 -2 5 4 C -2 -4 -4 -5 12 -5 -5 -3 -3 -2 -6 -5 -5 -4 -3 0 -2 -8 0 -2 -3 -4 Q 0 1 1 2 -5 4 2 -1 3 -2 -2 1 -1 -5 0 -1 -1 -5 -4 -2 3 5 E 0 -1 1 3 -5 2 4 0 1 -2 -3 0 -2 -5 -1 0 0 -7 -4 -2 4 5 G 1 -3 0 1 -3 -1 0 5 -2 -3 -4 -2 -3 -5 0 1 0 -7 -5 -1 2 1 H -1 2 2 1 -3 3 1 -2 6 -2 -2 0 -1 -1 -3 0 -2 3 3 I -1 -2 -2 -2 -3 -2 5 2 -2 2 1 -2 -1 0 -5 -1 4 -1 -1 L -2 -3 -3 -4 -6 -2 -3 -4 -2 2 6 -3 4 2 -3 -3 -2 -2 -1 K -1 3 1 0 -5 1 0 -2 -3 5 0 -5 -1 0 0 -3 -4 -2 2 2 M -1 0 -2 -3 -5 -1 -2 -3 -2 2 4 0 6 0 -2 -2 -1 -4 -2 2 -1 0 生物信息学 F -3 -4 -3 -6 -4 -5 -5 -5 -2 1 2 -5 0 9 -5 -3 -3 0 7 -1 -3 -4 P 1 0 0 -1 -3 0 -1 0 0 -2 -3 -1 -2 -5 6 1 0 -6 -5 -1 1 1 S 1 0 0 -1 0 1 -1 -1 -3 0 -2 -3 1 2 1 -2 -3 -1 2 1 T 1 -1 0 0 -2 -1 0 0 -1 0 -2 0 -1 -3 0 1 3 -5 -3 0 2 1 W -6 2 -4 -7 -8 -5 -7 -7 -3 -5 -2 -3 -4 0 -6 -2 -5 17 0 -6 -4 -4 Y -3 -4 -2 -4 0 -4 -4 -5 0 -1 -1 -4 -2 7 -5 -3 -3 0 10 -2 -2 -3 V 0 -2 -2 -2 -1 -2 4 2 -2 2 -1 -1 -1 0 -6 -2 4 0 0 B 2 1 4 5 -3 3 4 2 3 -1 -2 2 -1 -3 1 2 2 -4 -2 0 6 5 Z 1 2 3 4 -4 5 5 1 3 -1 -1 2 0 -4 1 1 1 -4 -3 0 5 6 83


BLOSUM 62 生物信息学 85


 Similarity vs homology
Similarity vs homology Compairson test
Compairson test Sequence identity vs similarity
Sequence identity vs similarity Sequence identity vs similarity
Sequence identity vs similarity Octopus sea star and grasshopper analogous or homologous
Octopus sea star and grasshopper analogous or homologous Homology vs homoplasy
Homology vs homoplasy Developmental homology
Developmental homology Homology modelling steps
Homology modelling steps Probability by homology
Probability by homology Homology modelling steps
Homology modelling steps Developmental homology
Developmental homology Homology modelling steps
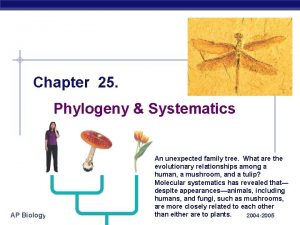
Homology modelling steps Ap biology phylogenetic tree
Ap biology phylogenetic tree Homology
Homology Derived homology
Derived homology Homology
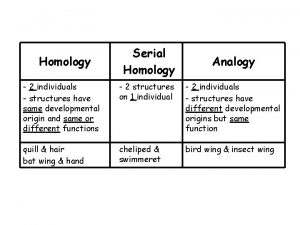
Homology Serially homologous
Serially homologous Homologous structures and analogous structures
Homologous structures and analogous structures Vestigial structures
Vestigial structures Homology is evidence of ______.
Homology is evidence of ______. Differentiate finite sequence from an infinite sequence
Differentiate finite sequence from an infinite sequence Amino acid nucleotide
Amino acid nucleotide Selection pseudocode
Selection pseudocode Convolutional sequence to sequence learning.
Convolutional sequence to sequence learning. Triangle similarity aa
Triangle similarity aa Comparison of characteristics of parents and offspring
Comparison of characteristics of parents and offspring Dilations and similarity in the coordinate plane
Dilations and similarity in the coordinate plane Chapter 7 similarity
Chapter 7 similarity Chapter 5 competitive rivalry and competitive dynamics
Chapter 5 competitive rivalry and competitive dynamics Dilations and similarity in the coordinate plane
Dilations and similarity in the coordinate plane Sss sas aa
Sss sas aa 7-2 similarity and transformations
7-2 similarity and transformations Similarity and dissimilarity in data mining
Similarity and dissimilarity in data mining Abcyz
Abcyz Lesson 5 triangle congruence and similarity
Lesson 5 triangle congruence and similarity Similarity between prokaryotic and eukaryotic cells
Similarity between prokaryotic and eukaryotic cells What is one similarity between ghana and mali
What is one similarity between ghana and mali Triangle similarity aa quiz
Triangle similarity aa quiz Similar images
Similar images 7-2 properties of proportions answers
7-2 properties of proportions answers Vector vs scalar quantities
Vector vs scalar quantities Projectile motion ball
Projectile motion ball Market commonality and resource similarity
Market commonality and resource similarity In metaphoric extension the novel stimulus shares
In metaphoric extension the novel stimulus shares Smc vs jaccard
Smc vs jaccard Uncut penis
Uncut penis Dilations and similarity
Dilations and similarity Standard cycle market example
Standard cycle market example Dilations and similarity in the coordinate plane
Dilations and similarity in the coordinate plane West side story romeo and juliet comparison
West side story romeo and juliet comparison Similarities and differences essay
Similarities and differences essay Osi vs tcp ip
Osi vs tcp ip Comparison and critique of osi and tcp/ip model
Comparison and critique of osi and tcp/ip model Ababababababababab
Ababababababababab Lesson 3 bisectors in triangles
Lesson 3 bisectors in triangles Similarity heuristic
Similarity heuristic Maze isosceles and equilateral triangles answer key
Maze isosceles and equilateral triangles answer key Side side side similarity postulate
Side side side similarity postulate Similarity flooding
Similarity flooding Similar images
Similar images Similarity of triangles
Similarity of triangles Orthogonal transformation
Orthogonal transformation The similarity of ending sounds existing between two words
The similarity of ending sounds existing between two words Earth similarity index
Earth similarity index 7-4 similarity in right triangles
7-4 similarity in right triangles Moss similarity
Moss similarity Narrow transcription
Narrow transcription Identity vs similarity protein
Identity vs similarity protein Similarity heuristic
Similarity heuristic Gestalt aesthetics
Gestalt aesthetics 7-3 assignment similar triangles
7-3 assignment similar triangles 9-4 similarity in right triangles
9-4 similarity in right triangles Geometry chapter 7
Geometry chapter 7 Sas similarity theorem
Sas similarity theorem Sas similarity theorem example
Sas similarity theorem example Aa postulate
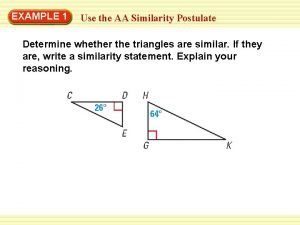
Aa postulate Postulate
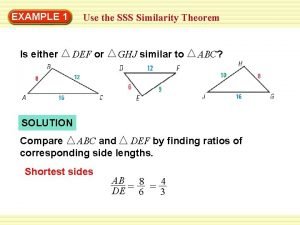
Postulate Sss similarity theorem
Sss similarity theorem Lnxn
Lnxn Chapter 7 similarity chapter test form a answer key
Chapter 7 similarity chapter test form a answer key Projected cognitive similarity is the tendency to
Projected cognitive similarity is the tendency to Ratios and proportions geometry
Ratios and proportions geometry 9-6 dilations
9-6 dilations 9-4 similarity in right triangles
9-4 similarity in right triangles 8-1 similarity in right triangles answer key
8-1 similarity in right triangles answer key 8-1 similarity in right triangles
8-1 similarity in right triangles 7-4 lesson quiz geometry
7-4 lesson quiz geometry Triangle similarity aa sss sas worksheet answers
Triangle similarity aa sss sas worksheet answers