Mutation Transposition and Repair A Gene Mutation 1


























- Slides: 49

Mutation, Transposition, and Repair

A. Gene Mutation 1. Mutations are Classified in Different Ways:

A. Gene Mutation 1. Mutations are Classified in Different Ways: a. By Cause: - spontaneous – just a ‘mistake’ (typically in replication) – assumed to be random

A. Gene Mutation 1. Mutations are Classified in Different Ways: a. By Cause: - spontaneous – just a ‘mistake’ (typically in replication) – assumed to be random - induced – caused by an external factor (mutagen); usually identified by increased rates of mutation above spontaneous levels in subpopulations exposed to the mutagen (radiation, chemicals)

A. Gene Mutation 1. Mutations are Classified in Different Ways: a. By Cause: b. By Location: - autosomal vs. sex-linked - somatic vs. germ-line (heritable)

A. Gene Mutation 1. Mutations are Classified in Different Ways: a. By Cause: b. By Location: c. By Type of Change: - substitution (“point mutation”)– the wrong base is inserted transition: purine for purine, etc. transversion: purine for pyrimidine, etc.

A. Gene Mutation 1. Mutations are Classified in Different Ways: a. By Cause: b. By Location: c. By Type of Change: - substitution (“point mutation”)– the wrong base is inserted transition: purine for purine, etc. transversion: purine for pyrimidine, etc. - substitutions may: change the amino acid (new codon): missense

A. Gene Mutation 1. Mutations are Classified in Different Ways: a. By Cause: b. By Location: c. By Type of Change: - substitution (“point mutation”)– the wrong base is inserted transition: purine for purine, etc. transversion: purine for pyrimidine, etc. - substitutions may: change the amino acid (new codon): missense not change the AA (redundancy): silent

A. Gene Mutation 1. Mutations are Classified in Different Ways: a. By Cause: b. By Location: c. By Type of Change: - substitution (“point mutation”)– the wrong base is inserted transition: purine for purine, etc. transversion: purine for pyrimidine, etc. - substitutions may: change the amino acid (new codon): missense not change the AA (redundancy): silent change to stop codon: nonsense

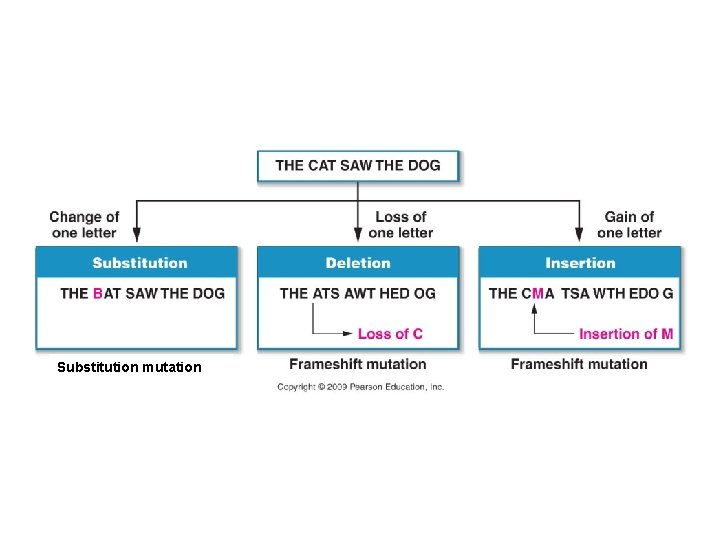
A. Gene Mutation 1. Mutations are Classified in Different Ways: a. By Cause: b. By Location: c. By Type of Change: - substitution (“point mutation”)– the wrong base is inserted - frameshift – bases are added or deleted, changing all codons downstream.

Substitution mutation

A. Gene Mutation 1. Mutations are Classified in Different Ways: a. By Cause: b. By Location: c. By Type of Change: d. By Effect on the Phenotype: - loss-of-function (null)

A. Gene Mutation 1. Mutations are Classified in Different Ways: a. By Cause: b. By Location: c. By Type of Change: d. By Effect on the Phenotype: - loss-of-function (null) - gain-of-function (enhanced or new function)

A. Gene Mutation 1. Mutations are Classified in Different Ways: a. By Cause: b. By Location: c. By Type of Change: d. By Effect on the Phenotype: - loss-of-function (null) - gain-of-function - neutral (change is not in a gene, or it is silent)

A. Gene Mutation 1. Mutations are Classified in Different Ways: a. By Cause: b. By Location: c. By Type of Change: d. By Effect on the Phenotype: - loss-of-function (null) - gain-of-function - neutral (change is not in a gene, or it is silent) - biochemical (physiological), morphological, behavioral

A. Gene Mutation 1. Mutations are Classified in Different Ways: a. By Cause: b. By Location: c. By Type of Change: d. By Effect on the Phenotype: - loss-of-function (null) - gain-of-function - neutral (change is not in a gene, or it is silent) - biochemical (physiological), morphological, behavioral - lethal and conditional

A. Gene Mutation 1. Mutations are Classified in Different Ways: 2. The Rates of Spontaneous Mutations: a. Mutation rates are low – selection has favored organisms that can replicate their DNA with few errors.

A. Gene Mutation 1. Mutations are Classified in Different Ways: 2. The Rates of Spontaneous Mutations: a. Mutation rates are low – selection has favored organisms that can replicate their DNA with few errors. b. But rates do vary by several orders of magnitude between different types of organisms. In higher eukaryotes, mutation rates (10 -5 – 10 -6) are higher than in bacteria and viruses (10 -8).

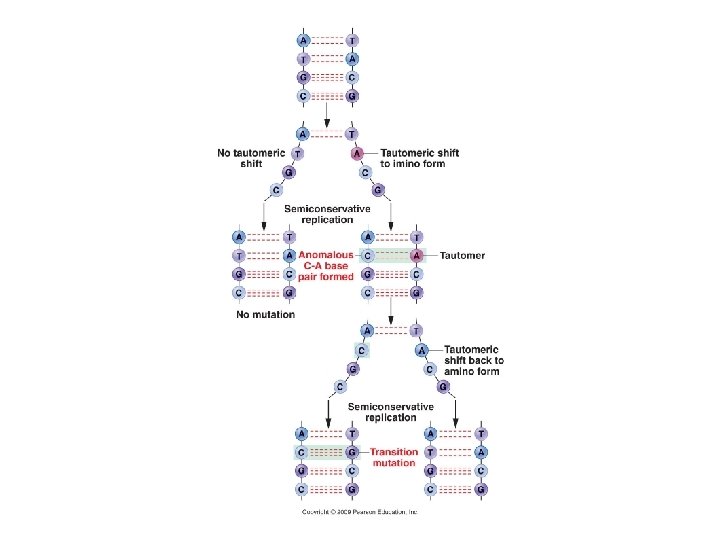
A. Gene Mutation 1. Mutations are Classified in Different Ways: 2. The Rates of Spontaneous Mutations: 3. How Spontaneous Mutations Occur: a. Substitutions: - truly random error in replication


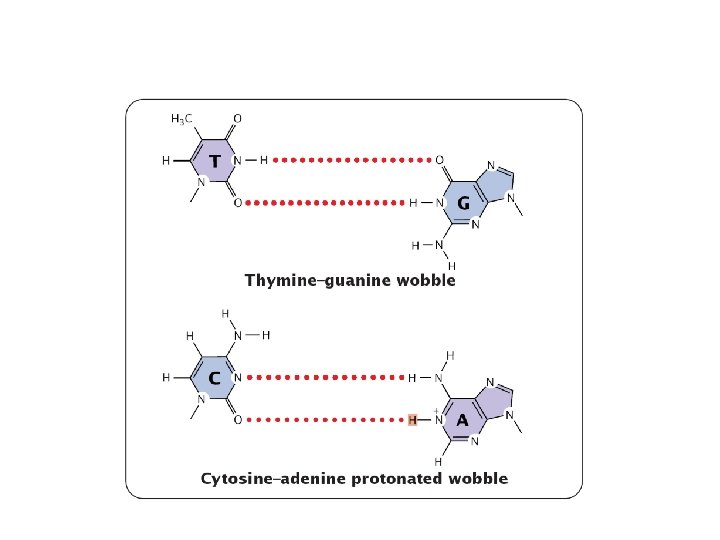
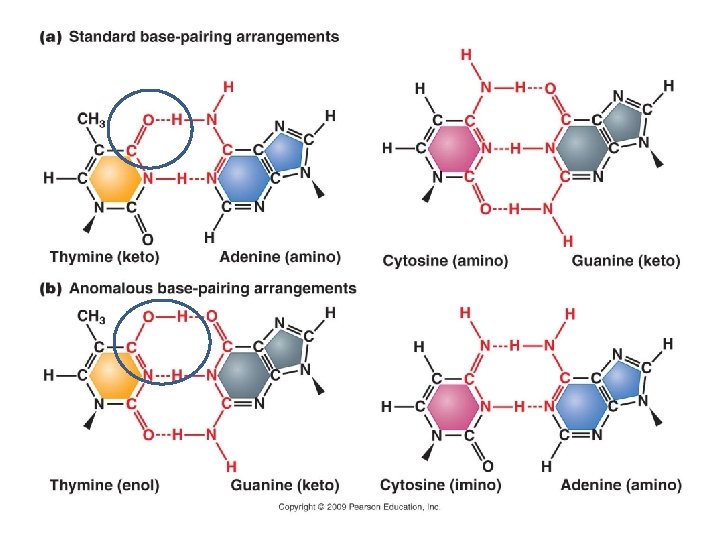
A. Gene Mutation 1. Mutations are Classified in Different Ways: 2. The Rates of Spontaneous Mutations: 3. How Spontaneous Mutations Occur: a. Substitutions: - truly random error in replication - tautomeric shift: All bases can exist in different forms. In the atypical form, they bind to different bases.



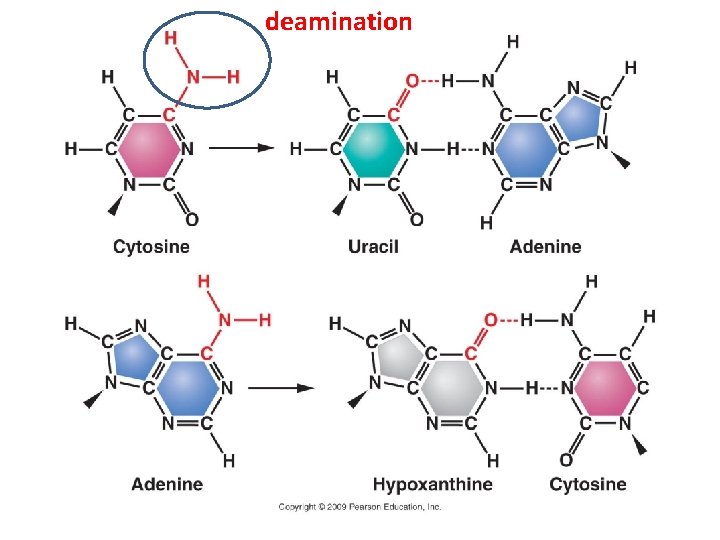
A. Gene Mutation 1. Mutations are Classified in Different Ways: 2. The Rates of Spontaneous Mutations: 3. How Spontaneous Mutations Occur: a. Substitutions: - truly random error in replication - tautomeric shift: (same base, but different pairing) - deamination of A and C cause mispairings

deamination

A. Gene Mutation 1. Mutations are Classified in Different Ways: 2. The Rates of Spontaneous Mutations: 3. How Spontaneous Mutations Occur: a. Substitutions: - truly random error in replication - tautomeric shift: (same base, but different pairing) - deamination of A and C cause mispairings - depurination: loss of A or G base in ds-DNA, and random replacement during replication.


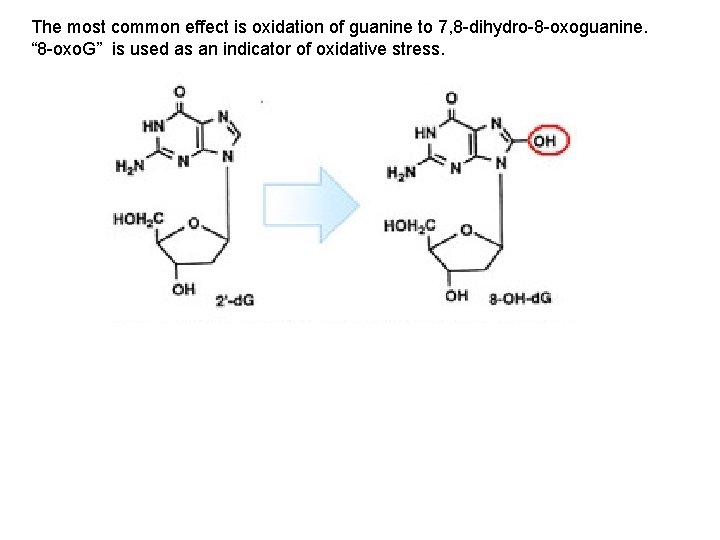
A. Gene Mutation 1. Mutations are Classified in Different Ways: 2. The Rates of Spontaneous Mutations: 3. How Spontaneous Mutations Occur: a. Substitutions: - truly random error in replication - tautomeric shift: (same base, but different pairing) - deamination of A and C cause mispairings - depurination: loss of A or G base in ds-DNA, and random replacement during replication. - oxidative damage to DNA due to normal metabolic production of oxidants, or “reactive oxygen species” (ROS) such as superoxides (O 2. -), hydroxl radicals (. OH), and hydrogen peroxide (H 2 O 2)

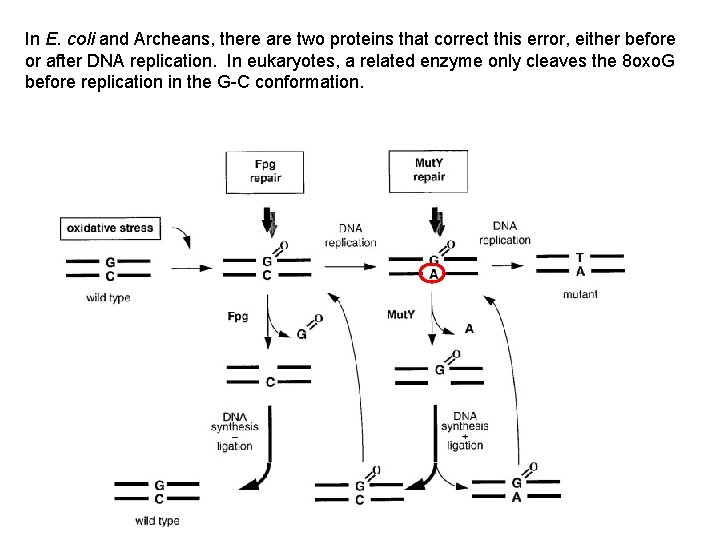
The most common effect is oxidation of guanine to 7, 8 -dihydro-8 -oxoguanine. “ 8 -oxo. G” is used as an indicator of oxidative stress.

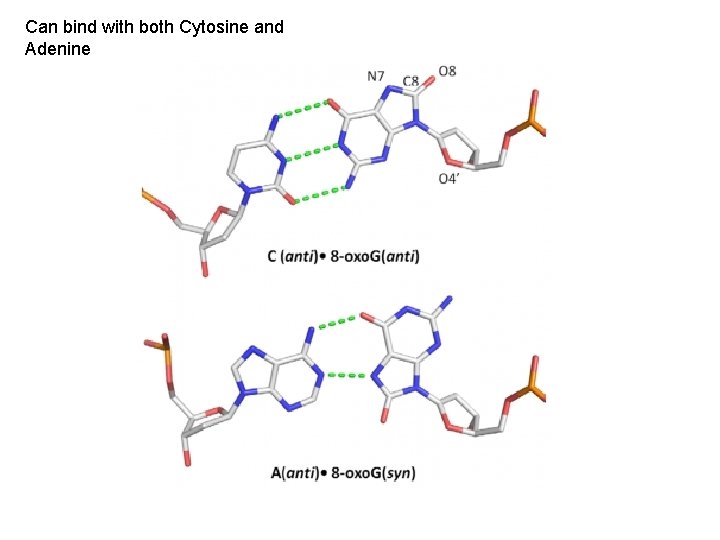
Can bind with both Cytosine and Adenine

In E. coli and Archeans, there are two proteins that correct this error, either before or after DNA replication. In eukaryotes, a related enzyme only cleaves the 8 oxo. G before replication in the G-C conformation.

Guanine is more susceptible to oxidation as the terminal G in a string of G’s (GGG) rather than as a single base in sequence. In this context, “G-C rich repeats” outside of genes may act as ‘oxidation pools’, soaking up the oxidative agents and protecting neighboring gene sequences. (Faucher, Doublié and Jia , 2012) http: //www. mdpi. com/1422 -0067/13/6/6711/htm

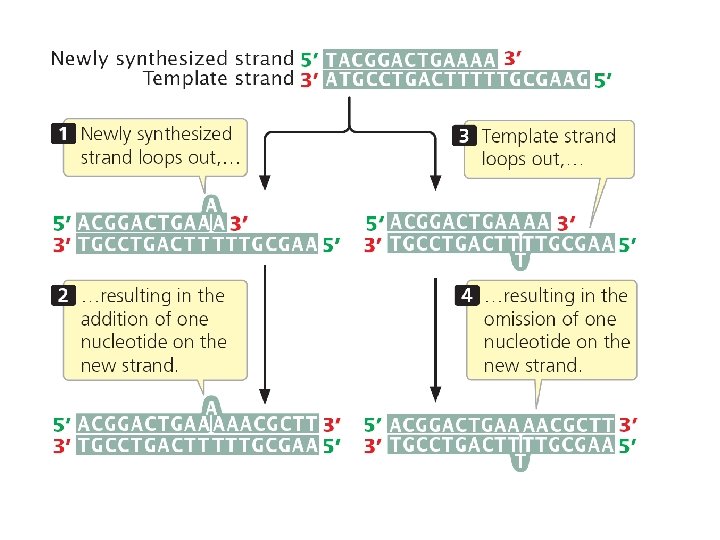
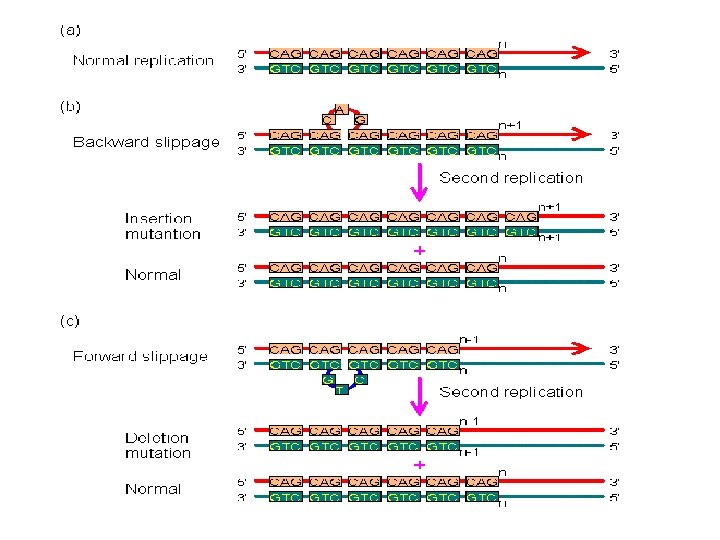
A. Gene Mutation 1. Mutations are Classified in Different Ways: 2. The Rates of Spontaneous Mutations: 3. How Spontaneous Mutations Occur: a. Substitutions: - truly random error in replication - tautomeric shift: (same base, but different pairing) - deamination of A and C cause mispairings - depurination: loss of A or G base in ds-DNA, and random replacement during replication. - oxidative damage b. Frameshifts: - replication slippage



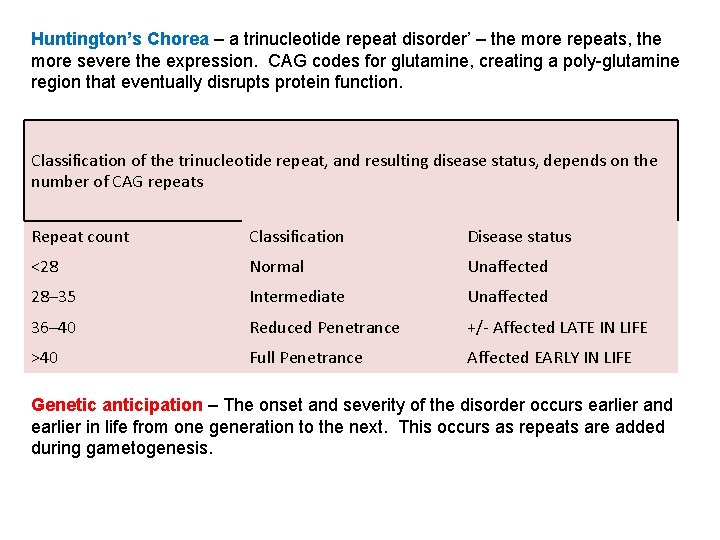
Huntington’s Chorea – a trinucleotide repeat disorder’ – the more repeats, the more severe the expression. CAG codes for glutamine, creating a poly-glutamine region that eventually disrupts protein function. Classification of the trinucleotide repeat, and resulting disease status, depends on the number of CAG repeats Repeat count Classification Disease status <28 Normal Unaffected 28– 35 Intermediate Unaffected 36– 40 Reduced Penetrance +/- Affected LATE IN LIFE >40 Full Penetrance Affected EARLY IN LIFE Genetic anticipation – The onset and severity of the disorder occurs earlier and earlier in life from one generation to the next. This occurs as repeats are added during gametogenesis.

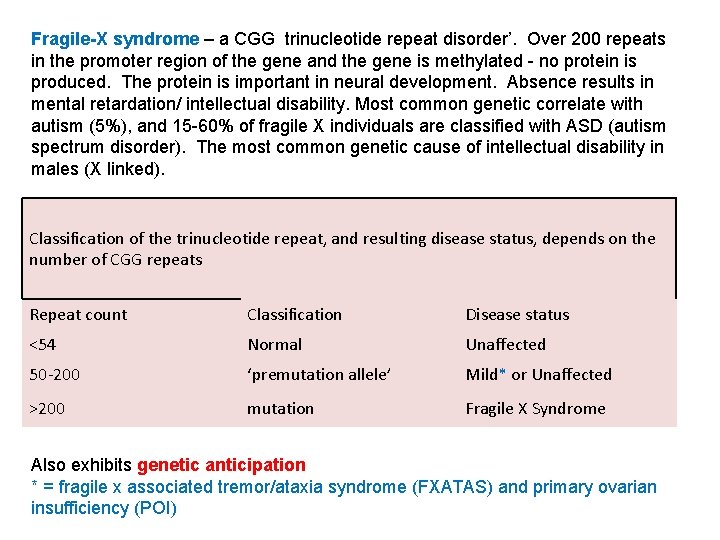
Fragile-X syndrome – a CGG trinucleotide repeat disorder’. Over 200 repeats in the promoter region of the gene and the gene is methylated - no protein is produced. The protein is important in neural development. Absence results in mental retardation/ intellectual disability. Most common genetic correlate with autism (5%), and 15 -60% of fragile X individuals are classified with ASD (autism spectrum disorder). The most common genetic cause of intellectual disability in males (X linked). Classification of the trinucleotide repeat, and resulting disease status, depends on the number of CGG repeats Repeat count Classification Disease status <54 Normal Unaffected 50 -200 ‘premutation allele’ Mild* or Unaffected >200 mutation Fragile X Syndrome Also exhibits genetic anticipation * = fragile x associated tremor/ataxia syndrome (FXATAS) and primary ovarian insufficiency (POI)

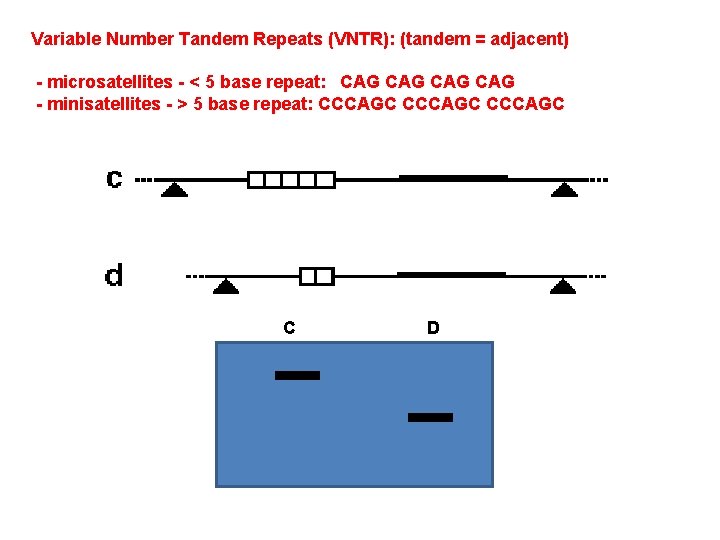
A. Gene Mutation 1. Mutations are Classified in Different Ways: 2. The Rates of Spontaneous Mutations: 3. How Spontaneous Mutations Occur: a. Substitutions: - truly random error in replication - tautomeric shift: (same base, but different pairing) - deamination of A and C cause mispairings - depurination: loss of A or G base in ds-DNA, and random replacement during replication. - oxidative damage b. Frameshifts: - replication slippage - most common where there are repeat sequences (like ‘tandem repeats’ of CGCGCGCGC…). - because errors are common, these are ‘hypermutable’ regions, and we differ at an individual level in the lengths of these sequences… often used for ‘DNA fingerprinting’

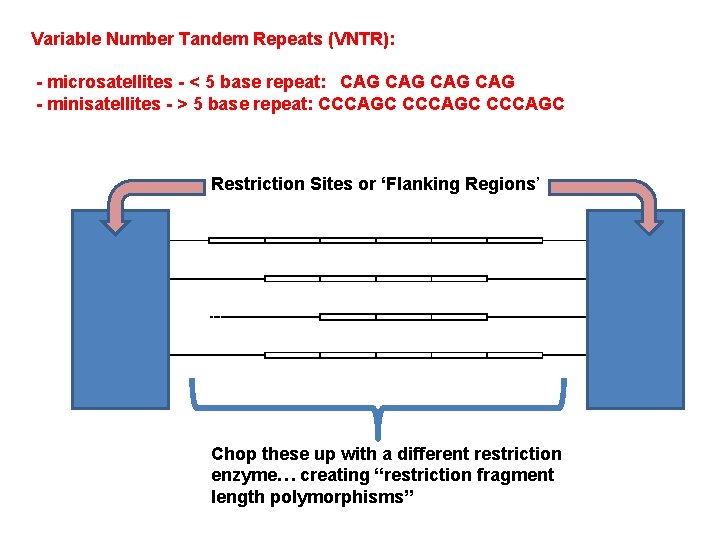
Variable Number Tandem Repeats (VNTR): (tandem = adjacent) - microsatellites - < 5 base repeat: CAG CAG - minisatellites - > 5 base repeat: CCCAGC C D

Variable Number Tandem Repeats (VNTR): - microsatellites - < 5 base repeat: CAG CAG - minisatellites - > 5 base repeat: CCCAGC Restriction Sites or ‘Flanking Regions’ Chop these up with a different restriction enzyme… creating “restriction fragment length polymorphisms”


A. Gene Mutation 1. Mutations are Classified in Different Ways: 2. The Rates of Spontaneous Mutations: 3. How Spontaneous Mutations Occur: a. Substitutions: - truly random error in replication - tautomeric shift: (same base, but different pairing) - deamination of A and C cause mispairings - depurination: loss of A or G base in ds-DNA, and random replacement during replication. - oxidative damage b. Frameshifts: - replication slippage - transposons – “jumping genes” can jump into an exon and turn a gene off jump into introns and affect splicing pattern – new gene “carry” a gene and multiply it through the genome

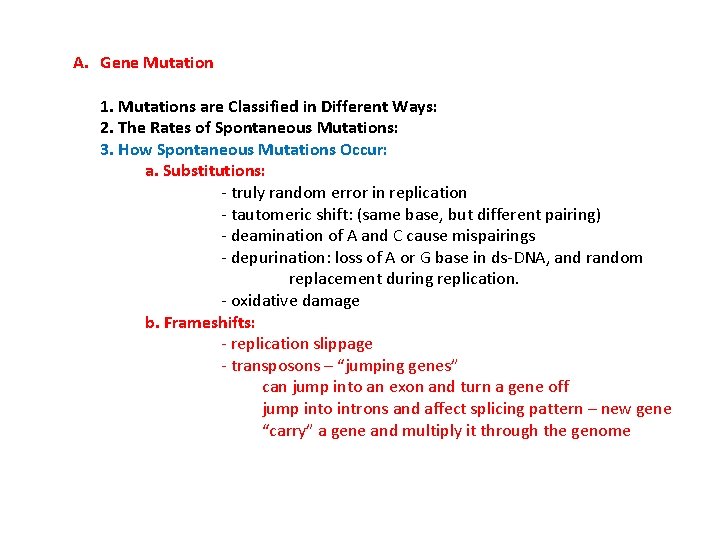
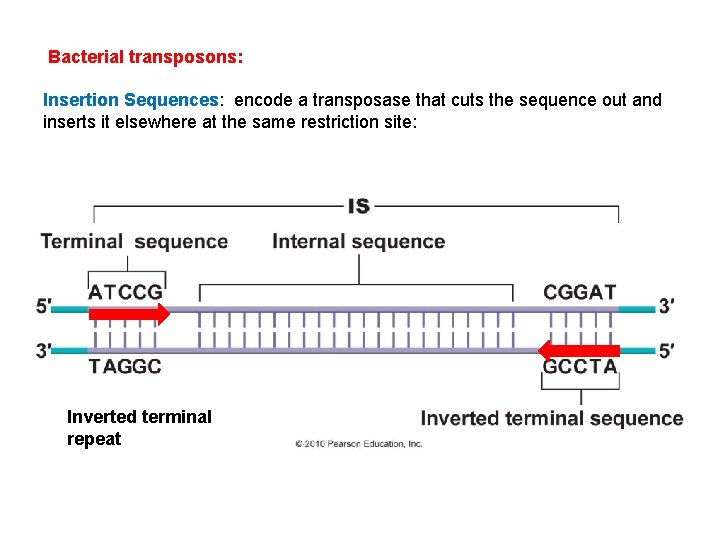
Bacterial transposons: Insertion Sequences: encode a transposase that cuts the sequence out and inserts it elsewhere at the same restriction site: Inverted terminal repeat

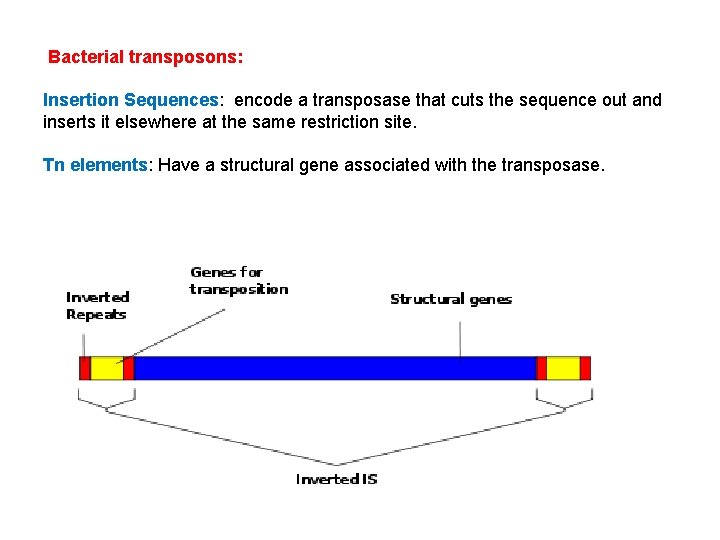
Bacterial transposons: Insertion Sequences: encode a transposase that cuts the sequence out and inserts it elsewhere at the same restriction site. Tn elements: Have a structural gene associated with the transposase.

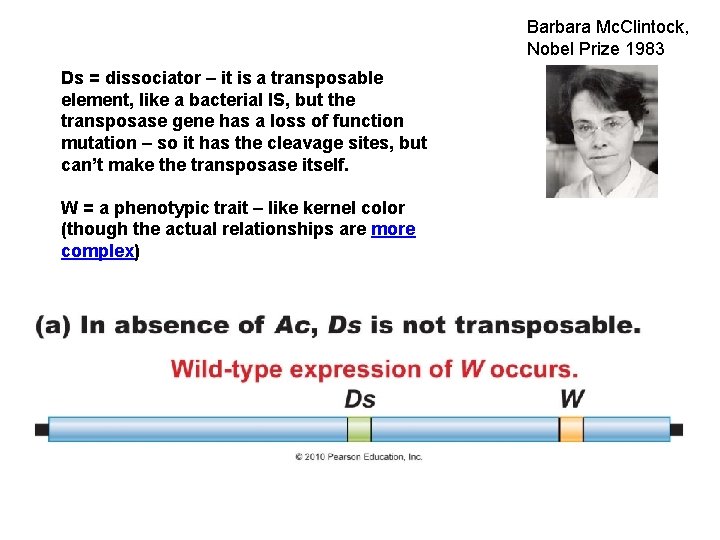
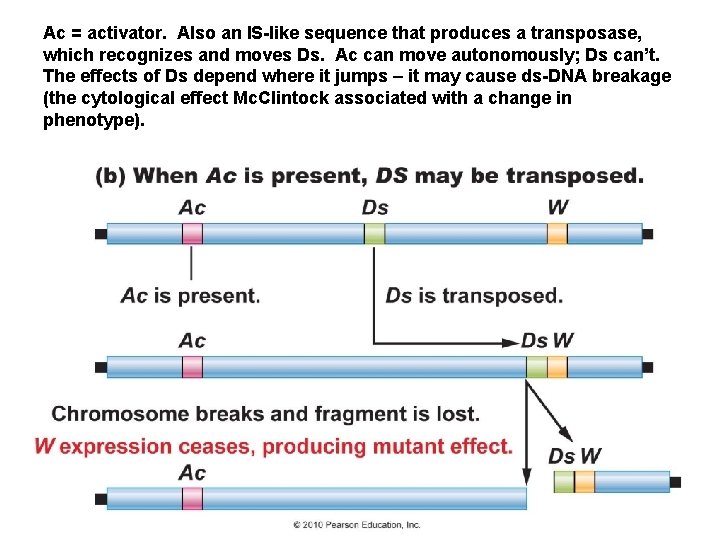
Barbara Mc. Clintock, Nobel Prize 1983 Ds = dissociator – it is a transposable element, like a bacterial IS, but the transposase gene has a loss of function mutation – so it has the cleavage sites, but can’t make the transposase itself. W = a phenotypic trait – like kernel color (though the actual relationships are more complex)

Ac = activator. Also an IS-like sequence that produces a transposase, which recognizes and moves Ds. Ac can move autonomously; Ds can’t. The effects of Ds depend where it jumps – it may cause ds-DNA breakage (the cytological effect Mc. Clintock associated with a change in phenotype).

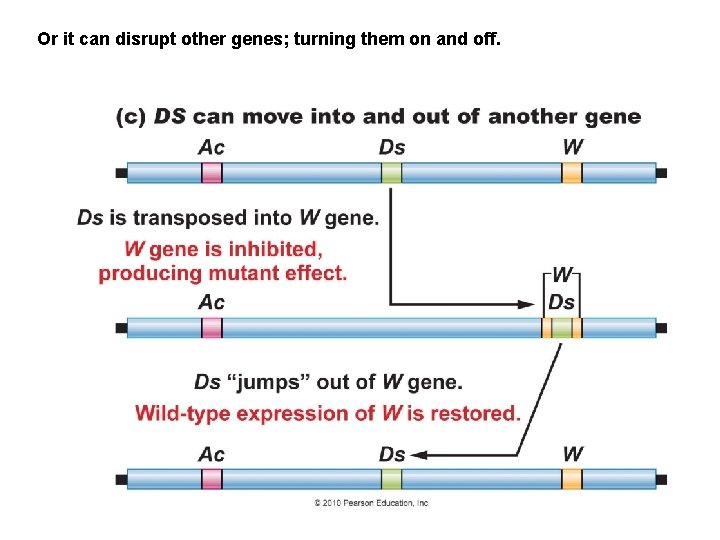
Or it can disrupt other genes; turning them on and off.

Humans: Long and Short Interspersed Elements (LINES and SINES) – 30% of the genome Other families of transposable elements = 11% Many are not mobile – their movement is repressed Diseases: - cases of hemophilia, Duchenne’s muscular dystrophy, and breast cancer have been identified that resulted from an insertion of a transposable element into a functional gene.

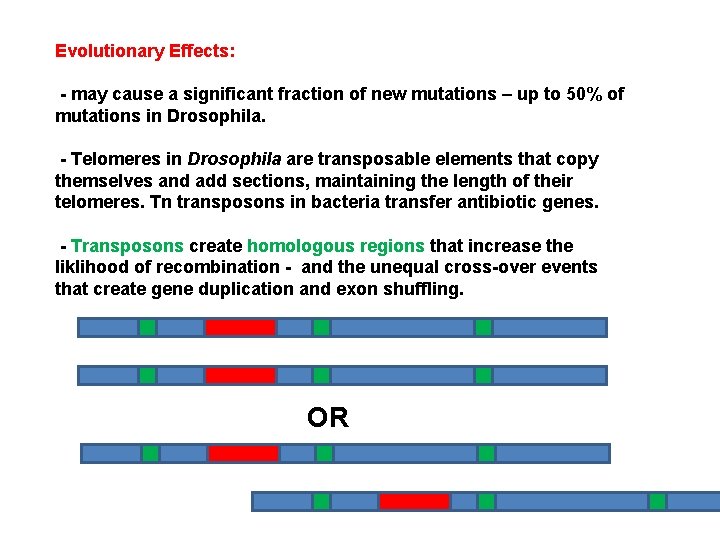
Evolutionary Effects: - may cause a significant fraction of new mutations – up to 50% of mutations in Drosophila. - Telomeres in Drosophila are transposable elements that copy themselves and add sections, maintaining the length of their telomeres. Tn transposons in bacteria transfer antibiotic genes. - Transposons create homologous regions that increase the liklihood of recombination - and the unequal cross-over events that create gene duplication and exon shuffling. OR