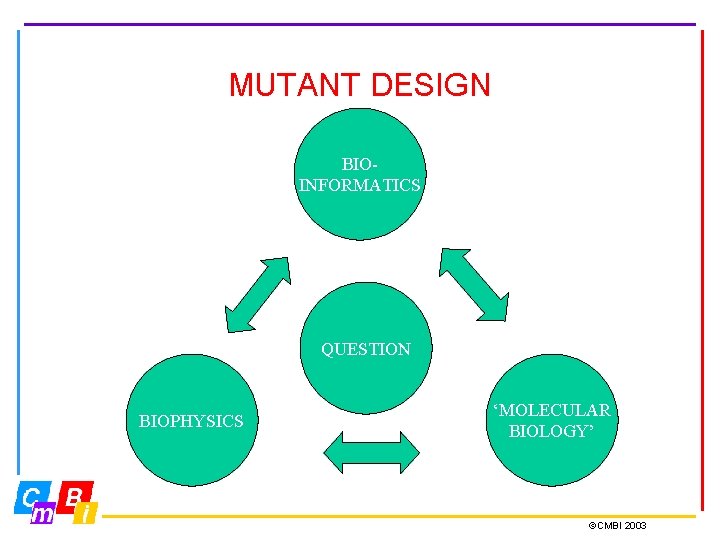
MUTANT DESIGN BIOINFORMATICS QUESTION BIOPHYSICS MOLECULAR BIOLOGY CMBI
MUTANT DESIGN BIOINFORMATICS QUESTION BIOPHYSICS ‘MOLECULAR BIOLOGY’ ©CMBI 2003
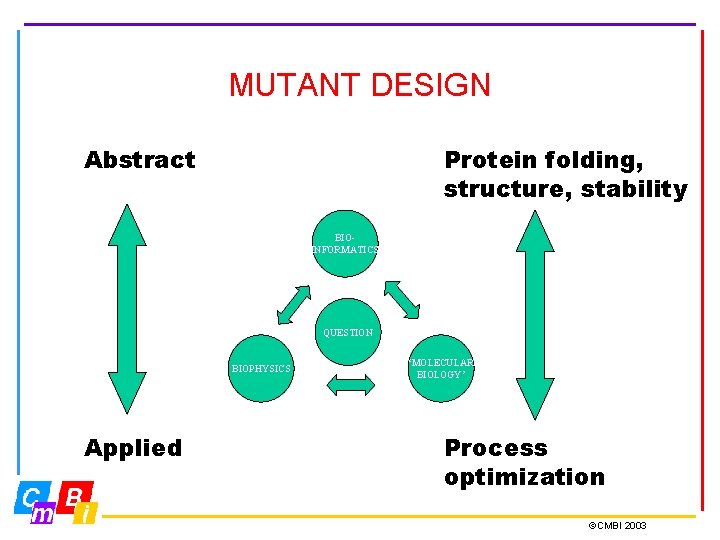
MUTANT DESIGN Abstract Protein folding, structure, stability BIOINFORMATICS QUESTION BIOPHYSICS Applied ‘MOLECULAR BIOLOGY’ Process optimization ©CMBI 2003

MUTANT DESIGN Three strong warnings and disclaimers: 1. I know nothing about MAKING mutants 2. Most times ‘evolutionary’ (that is grantwriting terminology for smart trial-anderror) beat design approaches. 3. Mutants are not always the best way to answer questions. Often good oldfashioned protein chemistry, spectroscopy, or even literature searches get you the answer more quickly. ©CMBI 2003
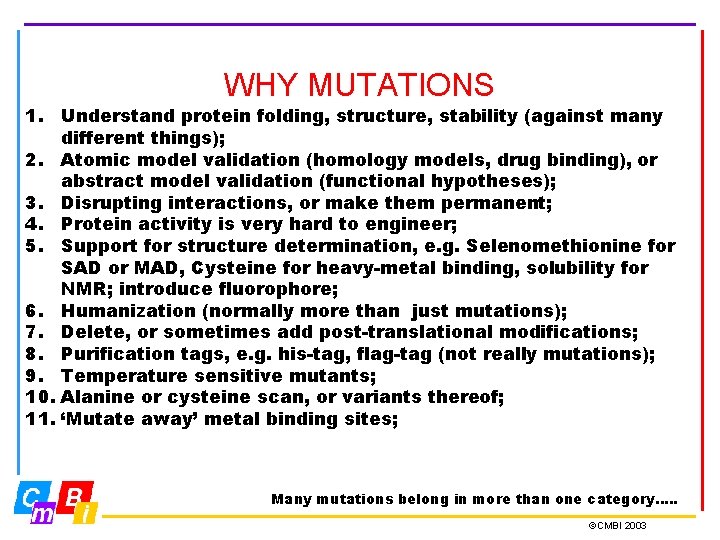
WHY MUTATIONS 1. Understand protein folding, structure, stability (against many different things); 2. Atomic model validation (homology models, drug binding), or abstract model validation (functional hypotheses); 3. Disrupting interactions, or make them permanent; 4. Protein activity is very hard to engineer; 5. Support for structure determination, e. g. Selenomethionine for SAD or MAD, Cysteine for heavy-metal binding, solubility for NMR; introduce fluorophore; 6. Humanization (normally more than just mutations); 7. Delete, or sometimes add post-translational modifications; 8. Purification tags, e. g. his-tag, flag-tag (not really mutations); 9. Temperature sensitive mutants; 10. Alanine or cysteine scan, or variants thereof; 11. ‘Mutate away’ metal binding sites; Many mutations belong in more than one category…. . ©CMBI 2003
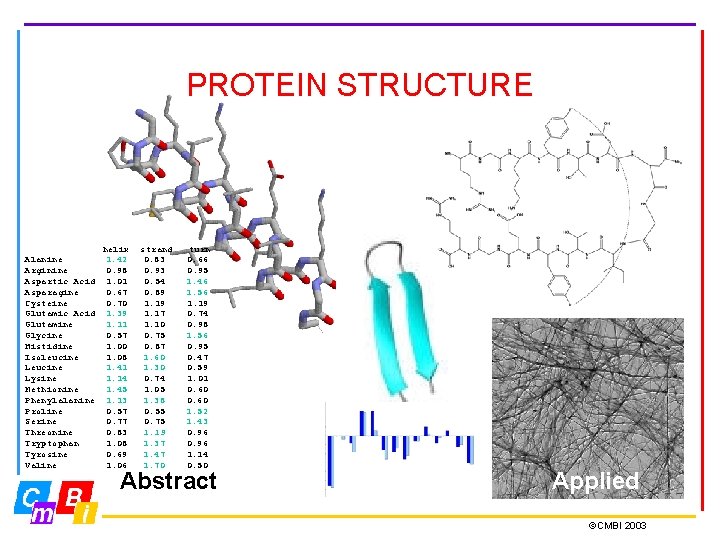
PROTEIN STRUCTURE Alanine Arginine Aspartic Acid Asparagine Cysteine Glutamic Acid Glutamine Glycine Histidine Isoleucine Lysine Methionine Phenylalanine Proline Serine Threonine Tryptophan Tyrosine Valine helix 1. 42 0. 98 1. 01 0. 67 0. 70 1. 39 1. 11 0. 57 1. 00 1. 08 1. 41 1. 14 1. 45 1. 13 0. 57 0. 77 0. 83 1. 08 0. 69 1. 06 strand 0. 83 0. 93 0. 54 0. 89 1. 17 1. 10 0. 75 0. 87 1. 60 1. 30 0. 74 1. 05 1. 38 0. 55 0. 75 1. 19 1. 37 1. 47 1. 70 turn 0. 66 0. 95 1. 46 1. 56 1. 19 0. 74 0. 98 1. 56 0. 95 0. 47 0. 59 1. 01 0. 60 1. 52 1. 43 0. 96 1. 14 0. 50 Abstract Applied ©CMBI 2003
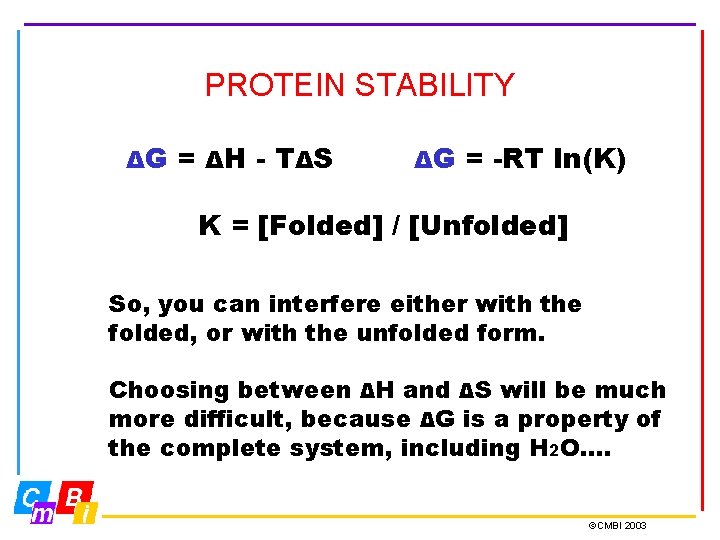
PROTEIN STABILITY ΔG = ΔH - T ΔS ΔG = -RT ln(K) K = [Folded] / [Unfolded] So, you can interfere either with the folded, or with the unfolded form. Choosing between ΔH and ΔS will be much more difficult, because ΔG is a property of the complete system, including H 2 O…. ©CMBI 2003
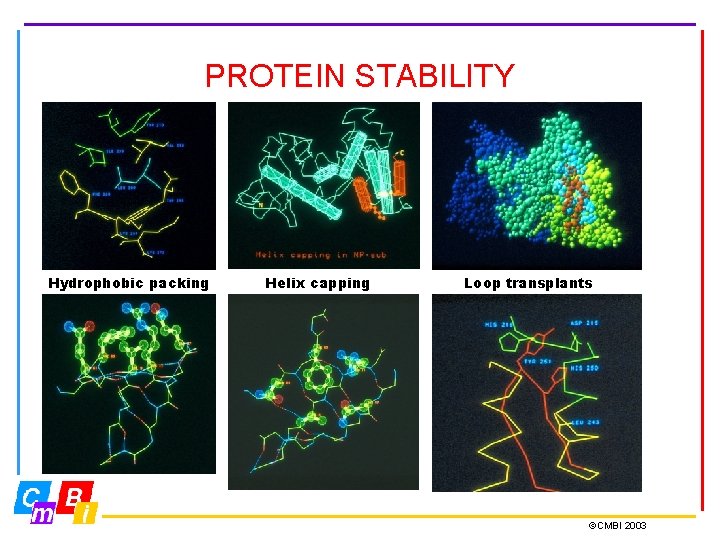
PROTEIN STABILITY Hydrophobic packing Helix capping Loop transplants ©CMBI 2003
PROTEIN STABILITY A whole series of tricks can be applied: Gly -> Any; Any -> Pro; Introduce hydrogen bonds; Hydrophobic packing; Cys-Cys bridges; Salt bridges; β-branched residues in β- strands; Pestering water from the core; etc. The main thing is that you should first know WHY the protein is unstable. Abstract: F U Applied: F LU I ©CMBI 2003
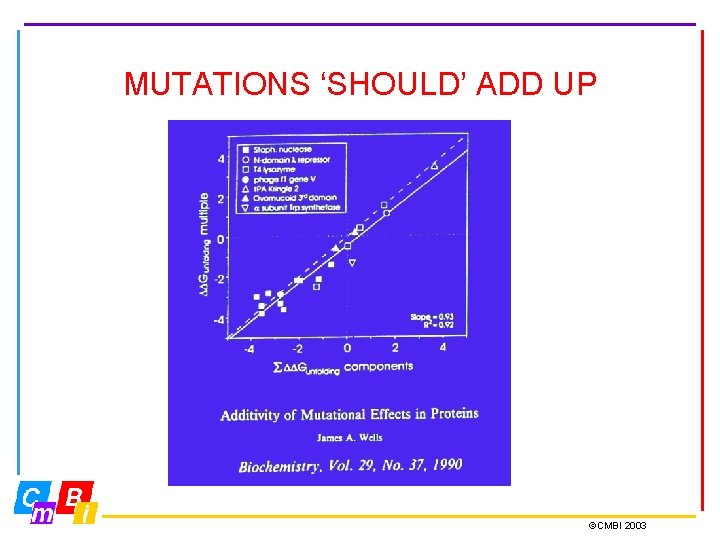
MUTATIONS ‘SHOULD’ ADD UP ©CMBI 2003
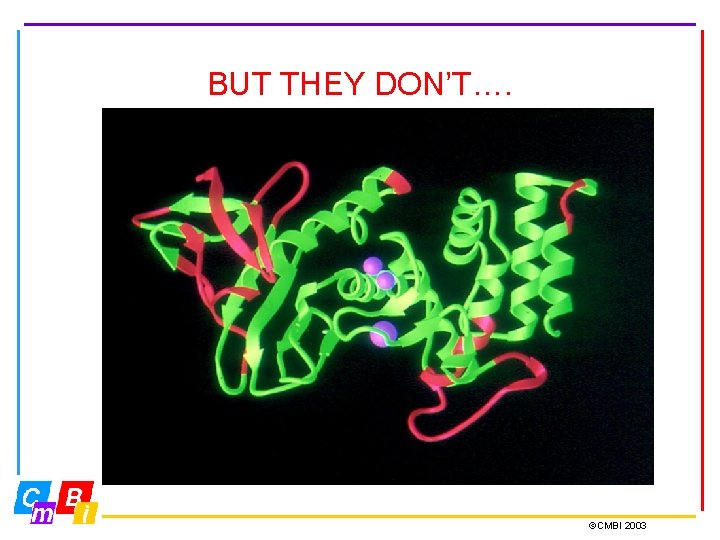
BUT THEY DON’T…. ©CMBI 2003
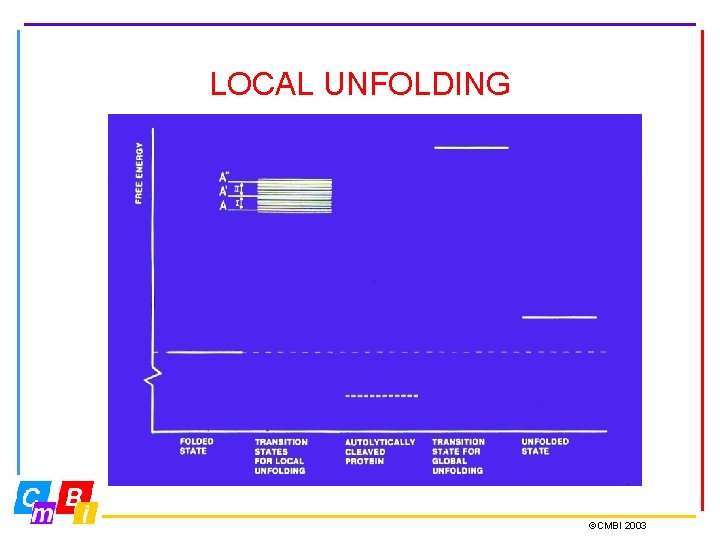
LOCAL UNFOLDING ©CMBI 2003
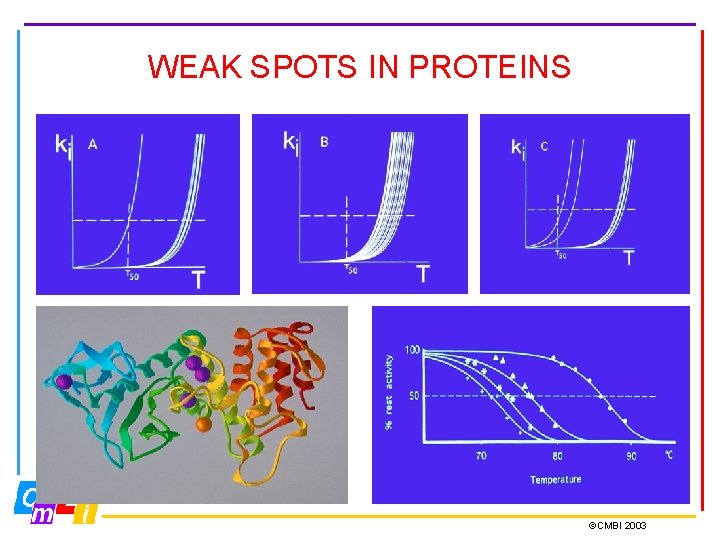
WEAK SPOTS IN PROTEINS ©CMBI 2003
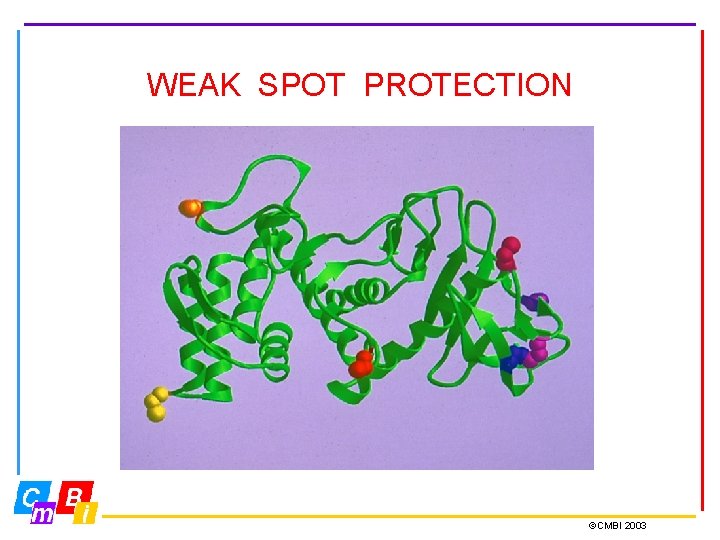
WEAK SPOT PROTECTION ©CMBI 2003

SUPPORT FOR EXPERIMENTS 1. 2. 3. 4. 5. 6. 7. 8. Selenomethionine for Xray; Solubility (i. e. for NMR); Tags for purification (His-tag, Flag-tag, etc); Addition or removal of post-translational modification sites; ‘Mutate away’ metal binding sites; Introduce fluorophore; Block binding, or make binding irreversible; Etcetera. ©CMBI 2003
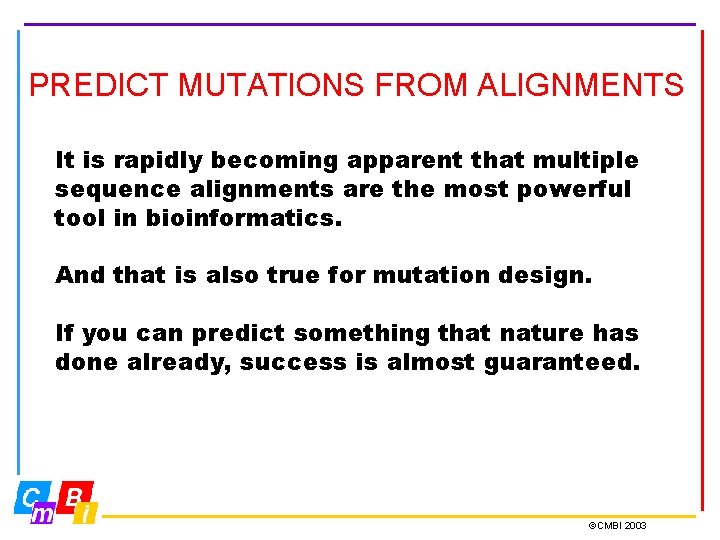
PREDICT MUTATIONS FROM ALIGNMENTS It is rapidly becoming apparent that multiple sequence alignments are the most powerful tool in bioinformatics. And that is also true for mutation design. If you can predict something that nature has done already, success is almost guaranteed. ©CMBI 2003
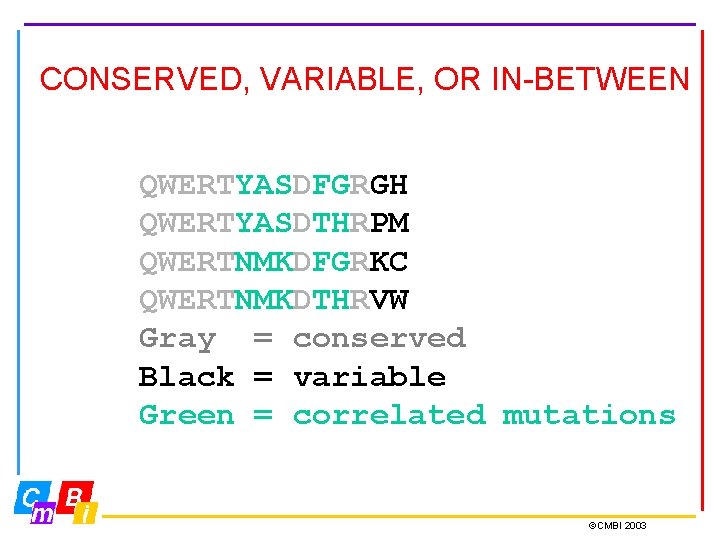
CONSERVED, VARIABLE, OR IN-BETWEEN QWERTYASDFGRGH QWERTYASDTHRPM QWERTNMKDFGRKC QWERTNMKDTHRVW Gray = conserved Black = variable Green = correlated mutations ©CMBI 2003
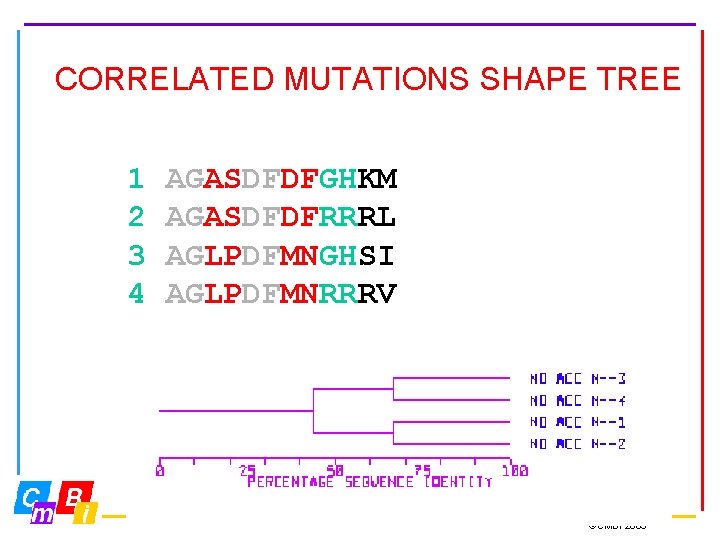
CORRELATED MUTATIONS SHAPE TREE 1 2 3 4 AGASDFDFGHKM AGASDFDFRRRL AGLPDFMNGHSI AGLPDFMNRRRV ©CMBI 2003
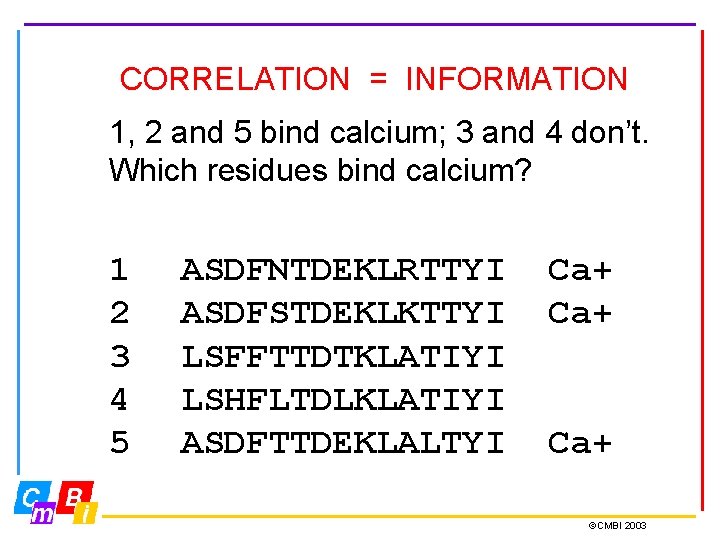
CORRELATION = INFORMATION 1, 2 and 5 bind calcium; 3 and 4 don’t. Which residues bind calcium? 1 2 3 4 5 ASDFNTDEKLRTTYI ASDFSTDEKLKTTYI LSFFTTDTKLATIYI LSHFLTDLKLATIYI ASDFTTDEKLALTYI Ca+ Ca+ ©CMBI 2003
- Slides: 18