Multivarietal genomic selection in French pig populations Cline



- Slides: 19

Multi-varietal genomic selection in French pig populations Céline Carillier-Jacquin Llibertat Tusell Palomero Juliette Riquet Marie-José Mercat Catherine Larzul INRA-Gen. Phy. SE, Toulouse, France 67 th EAAP meeting, 31 August 2016, Belfast, Northern Ireland . 01

Context • Small population size = lower GEBV accuracy • Nucleus from different breeding organizations > same breed but different populations • Including animals from a biggest population to small one = multivarietal genomic evaluation • Objective : look if multi-varietal evaluation could improve accuracy. 02

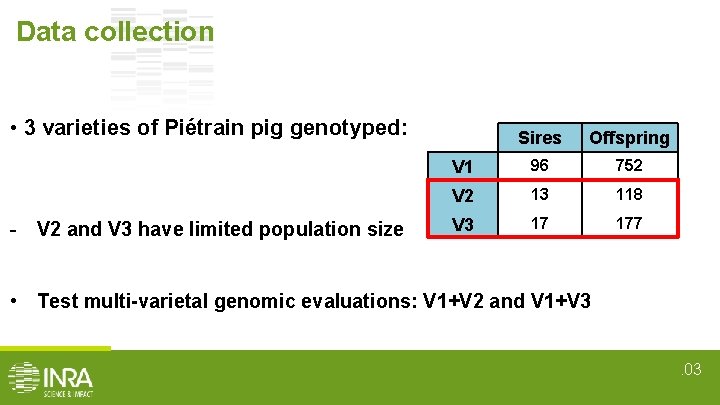
Data collection • 3 varieties of Piétrain pig genotyped: - V 2 and V 3 have limited population size Sires Offspring V 1 96 752 V 2 13 118 V 3 17 177 • Test multi-varietal genomic evaluations: V 1+V 2 and V 1+V 3. 03

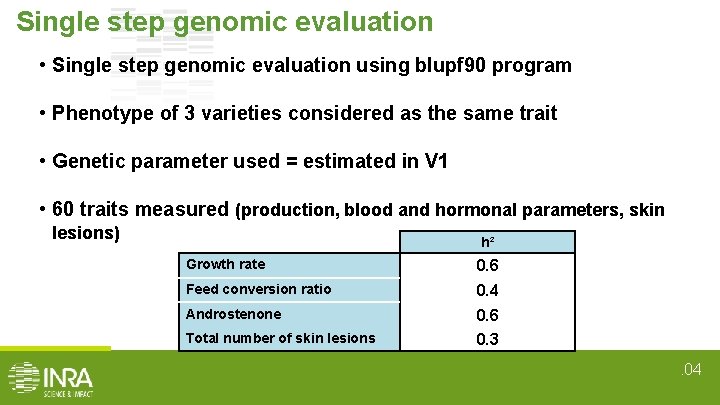
Single step genomic evaluation • Single step genomic evaluation using blupf 90 program • Phenotype of 3 varieties considered as the same trait • Genetic parameter used = estimated in V 1 • 60 traits measured (production, blood and hormonal parameters, skin lesions) h² Growth rate 0. 6 Feed conversion ratio 0. 4 Androstenone 0. 6 Total number of skin lesions 0. 3. 04

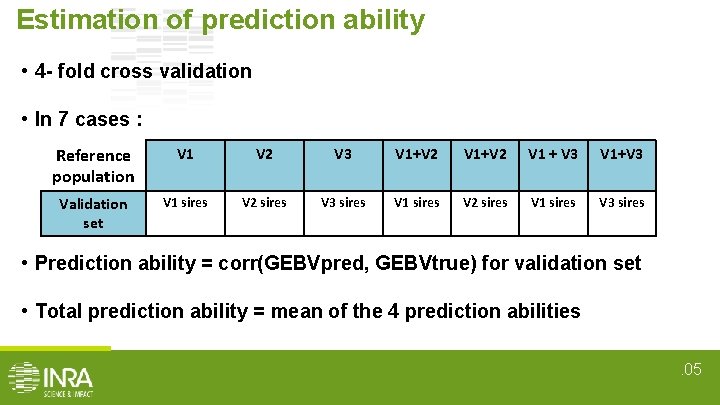
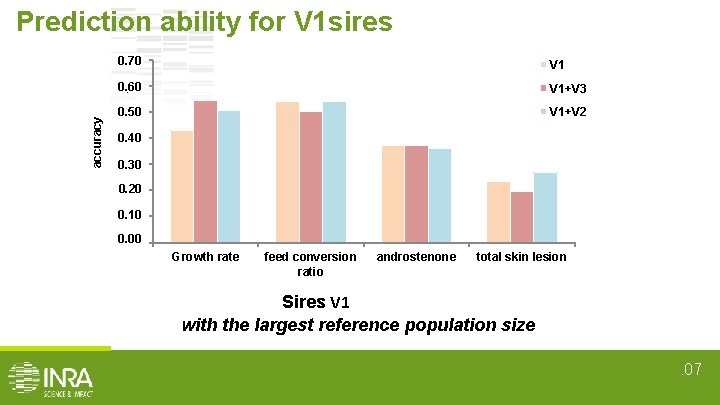
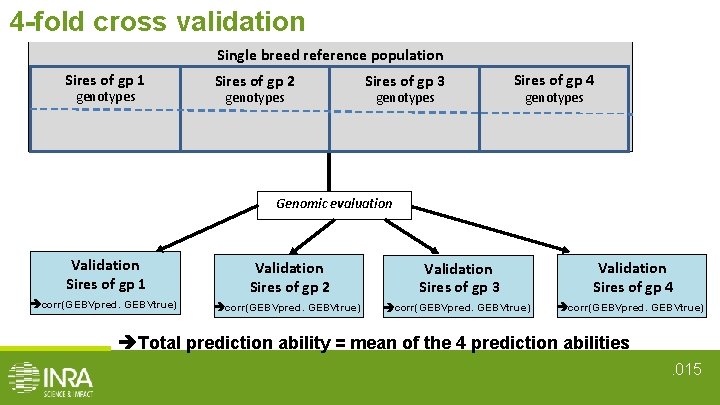
Estimation of prediction ability • 4 - fold cross validation • In 7 cases : Reference population V 1 V 2 V 3 V 1+V 2 V 1 + V 3 V 1+V 3 Validation set V 1 sires V 2 sires V 3 sires V 1 sires V 2 sires V 1 sires V 3 sires • Prediction ability = corr(GEBVpred, GEBVtrue) for validation set • Total prediction ability = mean of the 4 prediction abilities. 05

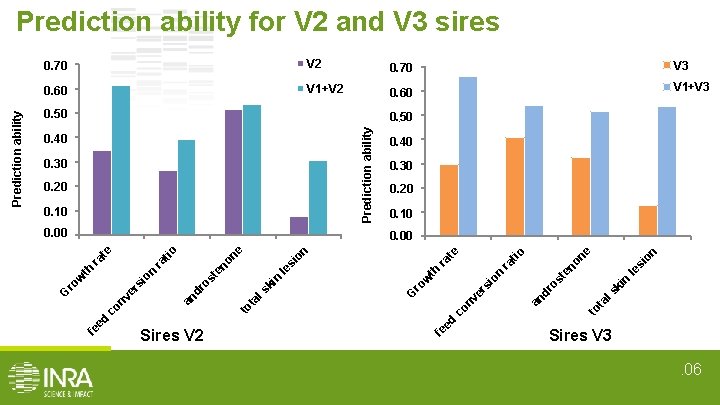
0, 70 0. 70 V 2 0, 70 0. 70 V 3 0, 60 0. 60 V 1+V 2 0, 60 0. 60 V 1+V 3 0, 50 0. 50 Prediction ability 0, 50 0, 40 0, 30 0. 20 0, 10 0. 00 0, 40 0, 30 0, 20 0, 10 0. 10 on ne le si no ki n ls to ta dr er nv co ed fe an si on w ro G os te ra ra th le si ki n ls to Sires V 2 tio te on ne no dr os te ta nv fe ed co an er ro w si on th ra ra tio te 0. 00 0, 00 G Prediction ability for V 2 and V 3 sires Sires V 3. 06

accuracy Prediction ability for V 1 sires 0. 70 0, 70 V 1 0. 60 0, 60 V 1+V 3 0. 50 0, 50 V 1+V 2 0. 40 0, 40 0. 30 0, 30 0. 20 0, 20 0. 10 0, 10 0. 00 0, 00 Growth rate feed conversion ratio androstenone total skin lesion Sires V 1 with the largest reference population size. 07

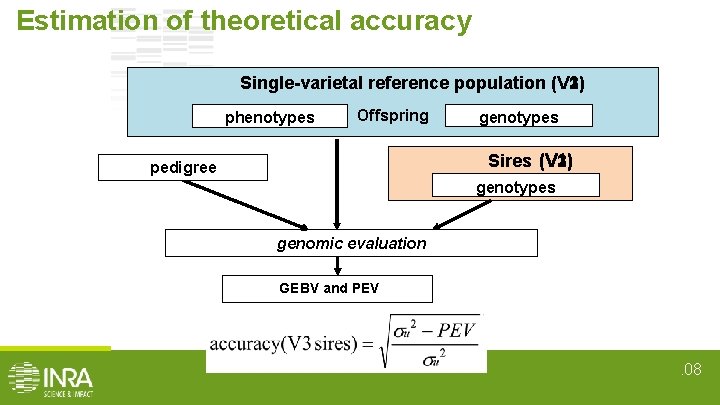
Estimation of theoretical accuracy Single-varietal reference population (V 2) (V 1) (V 3) phenotypes Offspring genotypes (V 1) (V 3) Sires (V 2) pedigree genotypes genomic evaluation GEBV and PEV . 08

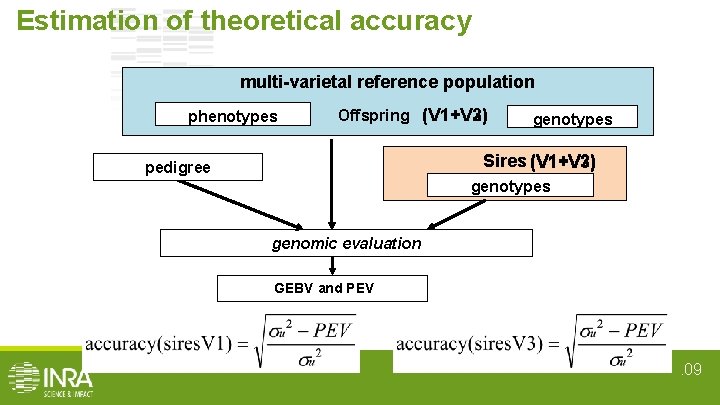
Estimation of theoretical accuracy multi-varietal reference population phenotypes Offspring (V 1+V 2) (V 1+V 3) genotypes Sires (V 1+V 3) (V 1+V 2) pedigree genotypes genomic evaluation GEBV and PEV . 09

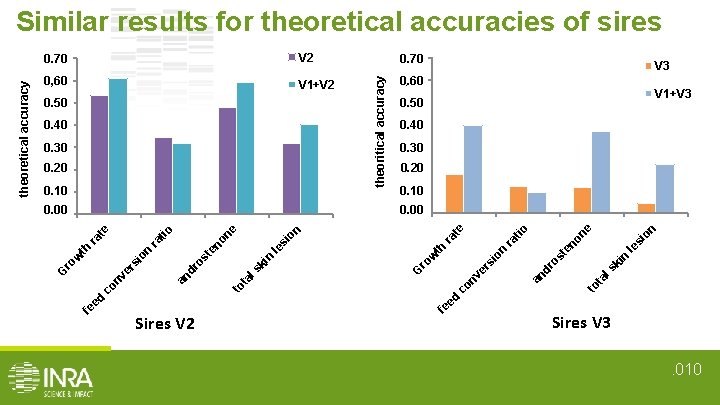
V 2 0, 70 0, 60 0. 60 V 1+V 2 0, 60 0. 50 0, 50 0. 40 0, 40 0. 30 0, 30 0. 20 0, 20 0. 10 0, 10 V 3 V 1+V 3 0, 50 0, 40 0, 30 0. 20 0, 20 0. 10 0, 10 on ne si le ki n ls ta to an er s fe ed co nv G dr io n os te ra no tio te th ro w le ki n ls to Sires V 2 ra on si no os te dr ed fe ta nv co an er ro w si on th ra ra tio ne 0. 00 0, 00 theoritical accuracy 0, 70 0. 70 G theoretical accuracy Similar results for theoretical accuracies of sires Sires V 3. 010

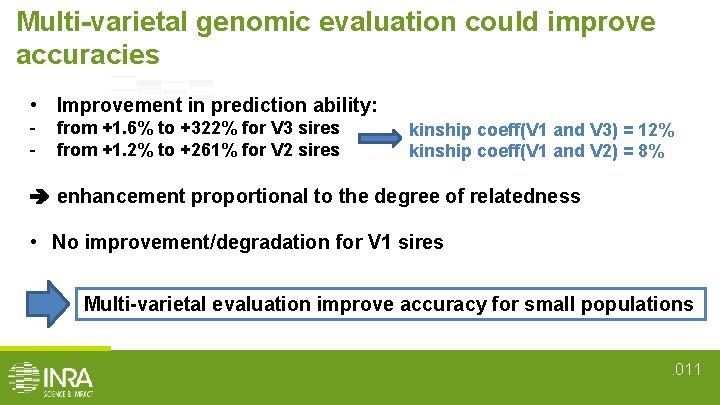
Multi-varietal genomic evaluation could improve accuracies • Improvement in prediction ability: - from +1. 6% to +322% for V 3 sires from +1. 2% to +261% for V 2 sires kinship coeff(V 1 and V 3) = 12% kinship coeff(V 1 and V 2) = 8% enhancement proportional to the degree of relatedness • No improvement/degradation for V 1 sires Multi-varietal evaluation improve accuracy for small populations. 011

Thank you for your attention! - Utopige fundings SELGEN - Bioporc (ADN, Choice Genetics France, Gene +, Nucleus) - IFIP and Le Rheu test station staff - Ignacy Misztal for blupf 90 suite of programs. 012

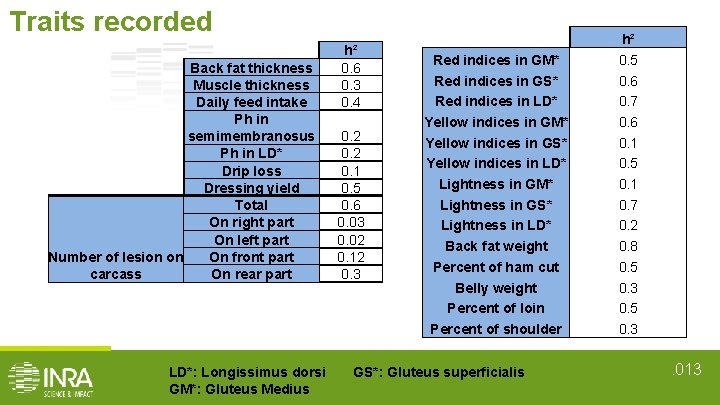
Traits recorded Back fat thickness Muscle thickness Daily feed intake Ph in semimembranosus Ph in LD* Drip loss Dressing yield Total On right part On left part On front part Number of lesion on carcass On rear part LD*: Longissimus dorsi GM*: Gluteus Medius h² 0. 6 0. 3 0. 4 0. 2 0. 1 0. 5 0. 6 0. 03 0. 02 0. 12 0. 3 Red indices in GM* Red indices in GS* Red indices in LD* Yellow indices in GM* Yellow indices in GS* Yellow indices in LD* Lightness in GM* Lightness in GS* Lightness in LD* Back fat weight Percent of ham cut Belly weight Percent of loin Percent of shoulder GS*: Gluteus superficialis h² 0. 5 0. 6 0. 7 0. 6 0. 1 0. 5 0. 1 0. 7 0. 2 0. 8 0. 5 0. 3 . 013

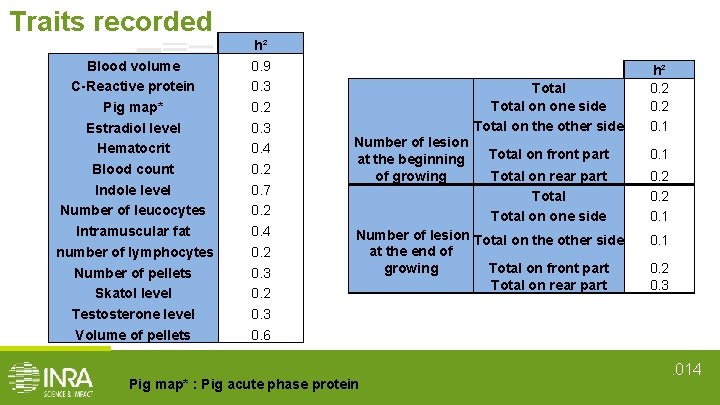
Traits recorded Blood volume C-Reactive protein Pig map* Estradiol level Hematocrit Blood count Indole level Number of leucocytes Intramuscular fat number of lymphocytes Number of pellets Skatol level Testosterone level Volume of pellets h² 0. 9 0. 3 0. 2 0. 3 0. 4 0. 2 0. 7 0. 2 0. 4 0. 2 0. 3 0. 6 Number of lesion at the beginning of growing Total on one side Total on the other side h² 0. 2 0. 1 Total on front part 0. 1 Total on rear part Total on one side 0. 2 0. 1 Number of lesion Total on the other side at the end of Total on front part growing Total on rear part Pig map* : Pig acute phase protein 0. 1 0. 2 0. 3 . 014

4 -fold cross validation Single breed reference population Sires of gp 1 Sires of gp 2 Sires of gp 3 Sires of gp 4 offsping genotypes genotypes + phenotypes Genomic evaluation Validation Sires of gp 1 Validation Sires of gp 2 Validation Sires of gp 3 Validation Sires of gp 4 corr(GEBVpred. GEBVtrue) Total prediction ability = mean of the 4 prediction abilities. 015

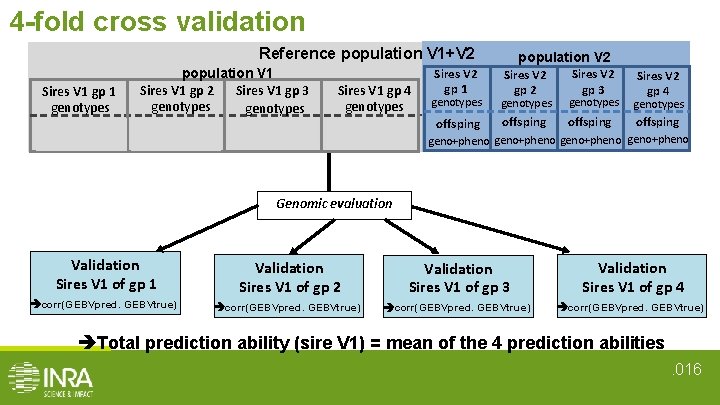
4 -fold cross validation Reference population V 1+V 2 Sires V 1 gp 1 genotypes offsping population V 1 Sires V 1 gp 3 Sires V 1 gp 2 genotypes offsping Sires V 1 gp 4 genotypes offsping geno + pheno Sires V 2 gp 1 population V 2 Sires V 2 gp 3 Sires V 2 gp 4 genotypes offsping geno+pheno Genomic evaluation Validation Sires V 1 of gp 1 Validation Sires V 1 of gp 2 Validation Sires V 1 of gp 3 Validation Sires V 1 of gp 4 corr(GEBVpred. GEBVtrue) Total prediction ability (sire V 1) = mean of the 4 prediction abilities. 016

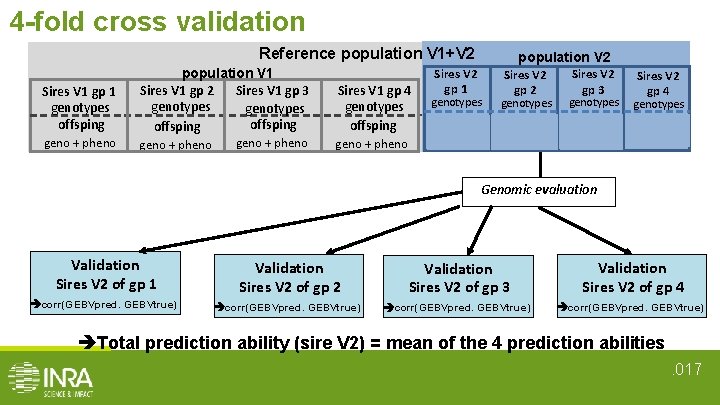
4 -fold cross validation Reference population V 1+V 2 Sires V 1 gp 1 genotypes offsping population V 1 Sires V 1 gp 3 Sires V 1 gp 2 genotypes offsping Sires V 1 gp 4 genotypes offsping geno + pheno Sires V 2 gp 1 population V 2 Sires V 2 gp 3 Sires V 2 gp 4 genotypes offsping geno+pheno Genomic evaluation Validation Sires V 2 of gp 1 Validation Sires V 2 of gp 2 Validation Sires V 2 of gp 3 Validation Sires V 2 of gp 4 corr(GEBVpred. GEBVtrue) Total prediction ability (sire V 2) = mean of the 4 prediction abilities. 017

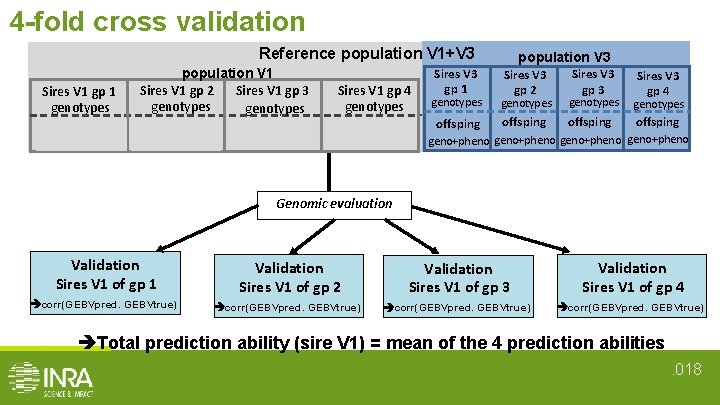
4 -fold cross validation Reference population V 1+V 3 Sires V 1 gp 1 genotypes offsping population V 1 Sires V 1 gp 3 Sires V 1 gp 2 genotypes offsping Sires V 1 gp 4 genotypes offsping geno + pheno Sires V 3 gp 1 population V 3 Sires V 3 gp 2 Sires V 3 gp 3 Sires V 3 gp 4 genotypes offsping geno+pheno Genomic evaluation Validation Sires V 1 of gp 1 Validation Sires V 1 of gp 2 Validation Sires V 1 of gp 3 Validation Sires V 1 of gp 4 corr(GEBVpred. GEBVtrue) Total prediction ability (sire V 1) = mean of the 4 prediction abilities. 018

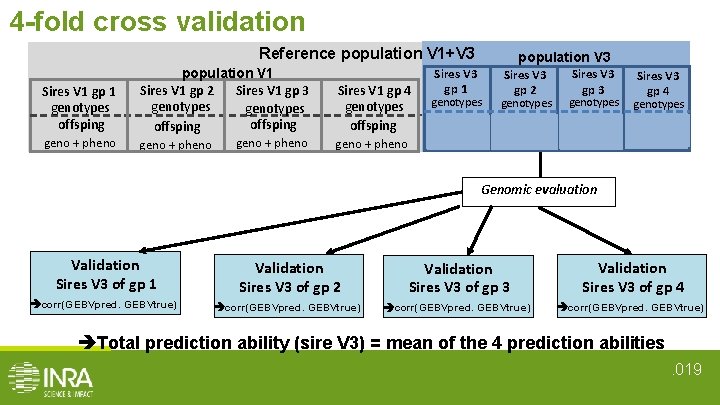
4 -fold cross validation Reference population V 1+V 3 Sires V 1 gp 1 genotypes offsping population V 1 Sires V 1 gp 3 Sires V 1 gp 2 genotypes offsping Sires V 1 gp 4 genotypes offsping geno + pheno Sires V 3 gp 1 population V 3 Sires V 3 gp 2 Sires V 3 gp 3 Sires V 3 gp 4 genotypes offsping geno+pheno Genomic evaluation Validation Sires V 3 of gp 1 Validation Sires V 3 of gp 2 Validation Sires V 3 of gp 3 Validation Sires V 3 of gp 4 corr(GEBVpred. GEBVtrue) Total prediction ability (sire V 3) = mean of the 4 prediction abilities. 019