MultiSite Genetic Analysis of 1151 Diffusion MRI Scans


- Slides: 18

Multi-Site Genetic Analysis of 1151 Diffusion MRI Scans from the ENIGMA–DTI Working Group Neda. Jahanshad@loni. ucla. edu Neda Jahanshad 1#, Peter Kochunov 2#, Emma Sprooten 3, 4, René C. Mandl 5, Thomas E. Nichols 6, 7, Laura Almasy 8, John Blangero 8, Rachel M. Brouwer 4, Joanne E. Curran 8, Greig I. de Zubicaray 9, Ravi Duggirala 8, Peter T. Fox 10, L. Elliot Hong 2, Bennett A. Landman 11, Nicholas G. Martin 12, Katie L. Mc. Mahon 13, Sarah E. Medland 12, Braxton D. Mitchell 14, Rene L. Olvera 10, Charles P. Peterson 8, John M. Starr 15, Jessika E. Sussmann 4, Arthur W. Toga 1, Joanna M. Wardlaw 15, Margaret J. Wright 12, Hilleke E. Hulshoff Pol 5, Mark E. Bastin 4, 15, Andrew M. Mc. Intosh 4, Ian J. Deary 15, Paul M. Thompson 1*and David C. Glahn 3 Joint work with UCLA, U. MD, U. Edinburgh, UMC Utrecht, U. Warwick, Vanderbilt, U. TX San Antonio, U. Queensland, QTIM, Yale

Meta analysis for genetic studies • A search for “meta-analysis” revealed 400+ articles in Nature Genetics in the last 10 years! – ~20%!! • What is meta-analysis? • Genetics - MA Important discoveries for: – AD/neurodegeneration – Psychiatric disorders – Behavior and cognition

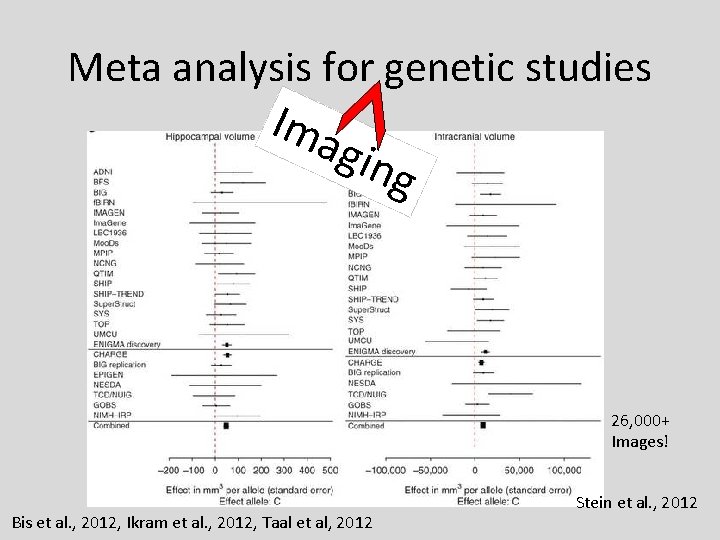
Meta analysis for genetic studies Ima gin g 26, 000+ Images! Bis et al. , 2012, Ikram et al. , 2012, Taal et al, 2012 Stein et al. , 2012

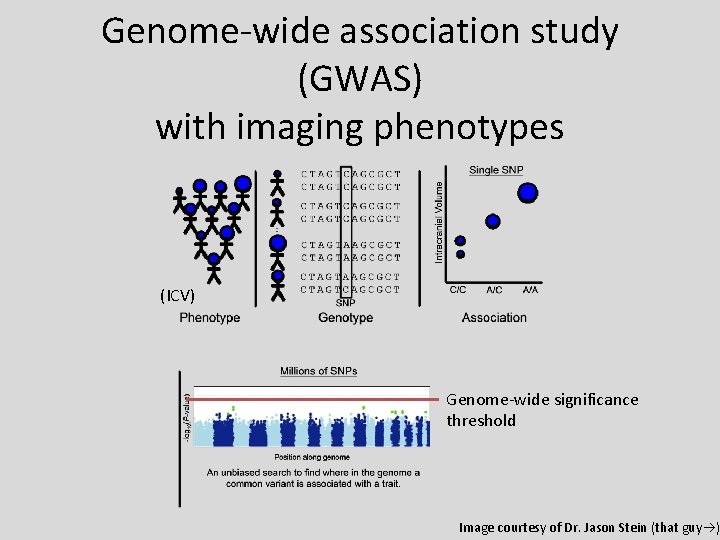
Genome-wide association study (GWAS) with imaging phenotypes (ICV) Genome-wide significance threshold Image courtesy of Dr. Jason Stein (that guy )

White Matter - Heritability, Risk Genes DTI

GWAS using DTI phenotypes • Single site studies of white matter integrity – Global measure of FA – General factor of FA • Top SNPs in candidate pathways • No “genome-wide” significant findings – Relatively small sample sizes – N~150, N~700

Multi-site DTI • Greatly increase the number of subjects – Genome wide significance? • Would a tract from one study be the same as a tract from another? • How reliable are the tracts in terms of imaging protocol and resolution • DTI reliability studies – Magnotta et al. , 2012 – Jahanshad et al. , 2010, 2012 – Zhan et al. , 2012 – Walker et al. , 2012 – Teipel et al. , 2011 • Planning studies, not merging existing data

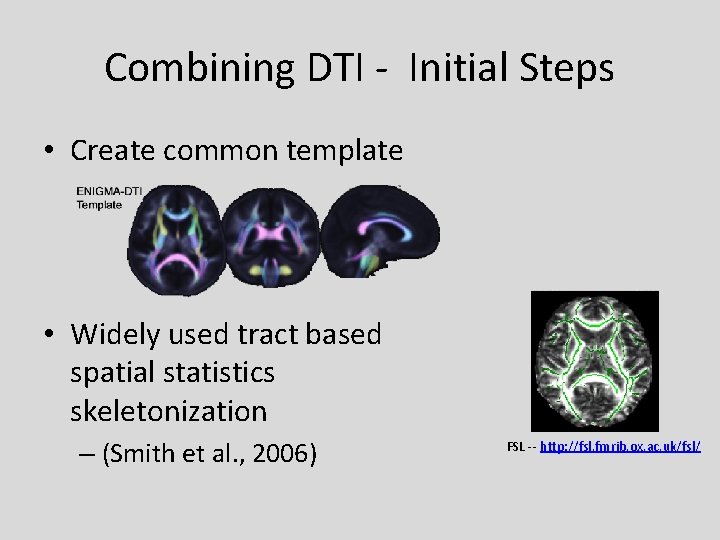
Combining DTI - Initial Steps • Create common template • Widely used tract based spatial statistics skeletonization – (Smith et al. , 2006) FSL -- http: //fsl. fmrib. ox. ac. uk/fsl/

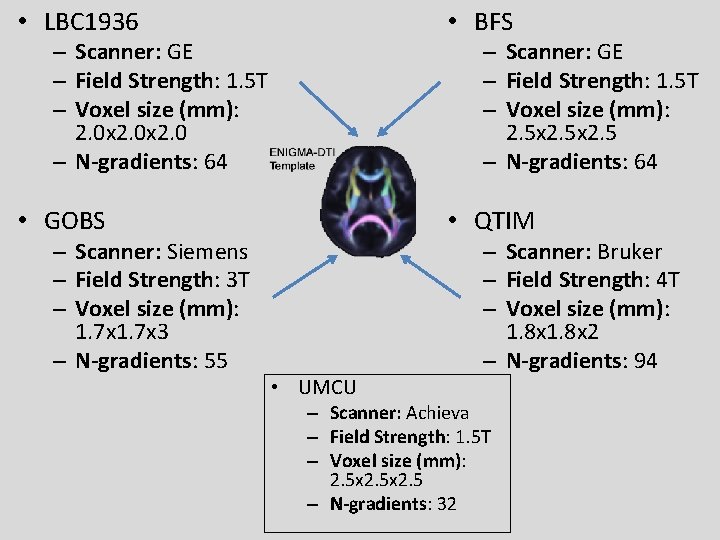
• LBC 1936 • BFS • GOBS • QTIM – Scanner: GE – Field Strength: 1. 5 T – Voxel size (mm): 2. 0 x 2. 0 – N-gradients: 64 – Scanner: Siemens – Field Strength: 3 T – Voxel size (mm): 1. 7 x 3 – N-gradients: 55 – Scanner: GE – Field Strength: 1. 5 T – Voxel size (mm): 2. 5 x 2. 5 – N-gradients: 64 • UMCU – Scanner: Bruker – Field Strength: 4 T – Voxel size (mm): 1. 8 x 2 – N-gradients: 94 – Scanner: Achieva – Field Strength: 1. 5 T – Voxel size (mm): 2. 5 x 2. 5 – N-gradients: 32

Multi-site DTI GWAS • Would tracts from different populations or datasets even be similarly heritable? – Blokland et al. , 2012 • Could they be used as endophenotypes for GWAS regardless of protocol?

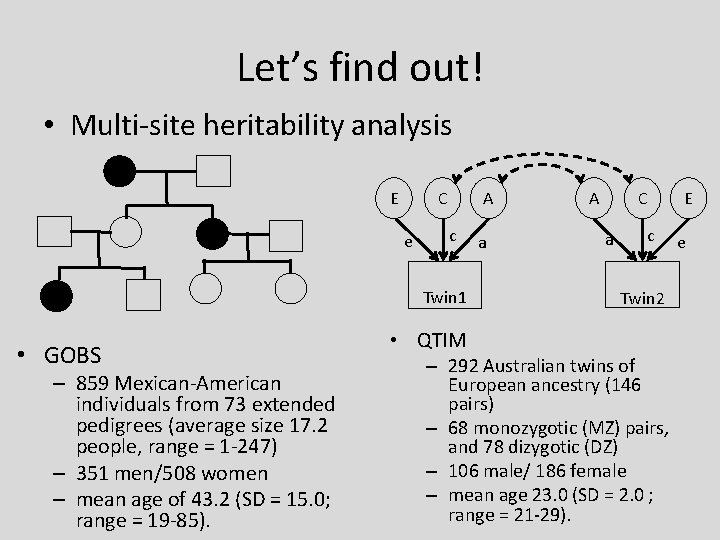
Let’s find out! • Multi-site heritability analysis A C E e c Twin 1 • GOBS – 859 Mexican-American individuals from 73 extended pedigrees (average size 17. 2 people, range = 1 -247) – 351 men/508 women – mean age of 43. 2 (SD = 15. 0; range = 19 -85). • QTIM a C A a c Twin 2 – 292 Australian twins of European ancestry (146 pairs) – 68 monozygotic (MZ) pairs, and 78 dizygotic (DZ) – 106 male/ 186 female – mean age 23. 0 (SD = 2. 0 ; range = 21 -29). E e

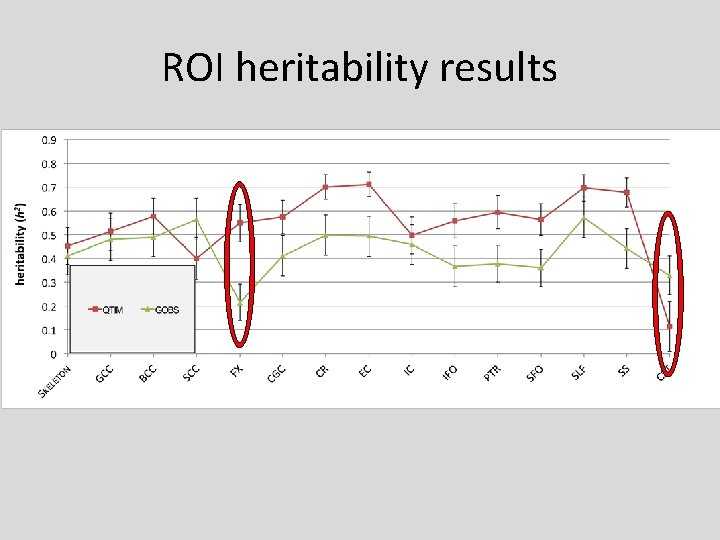
ROI heritability results

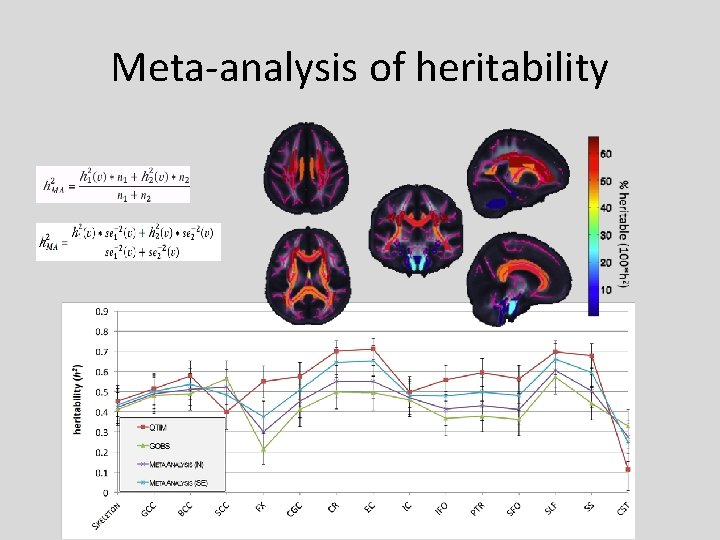
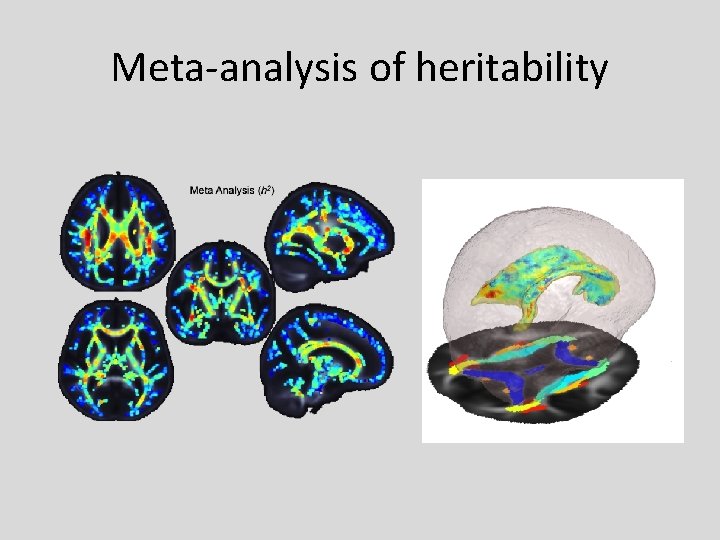
Meta-analysis of heritability

Meta-analysis of heritability

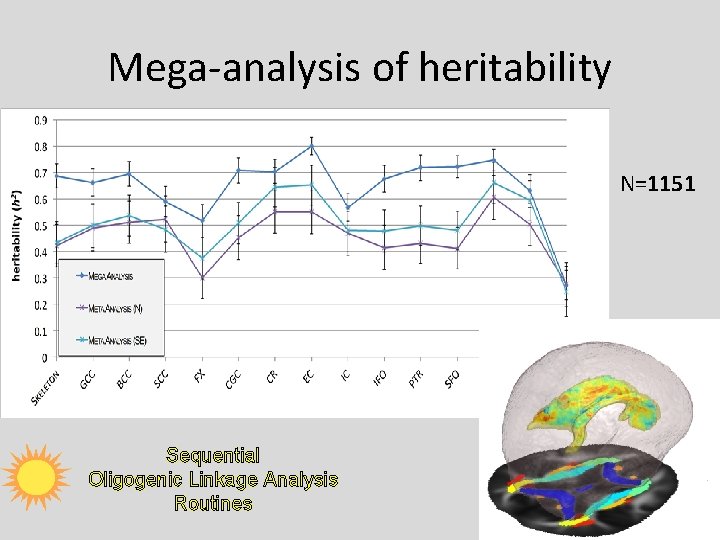
Mega-analysis of heritability N=1151 Sequential Oligogenic Linkage Analysis Routines

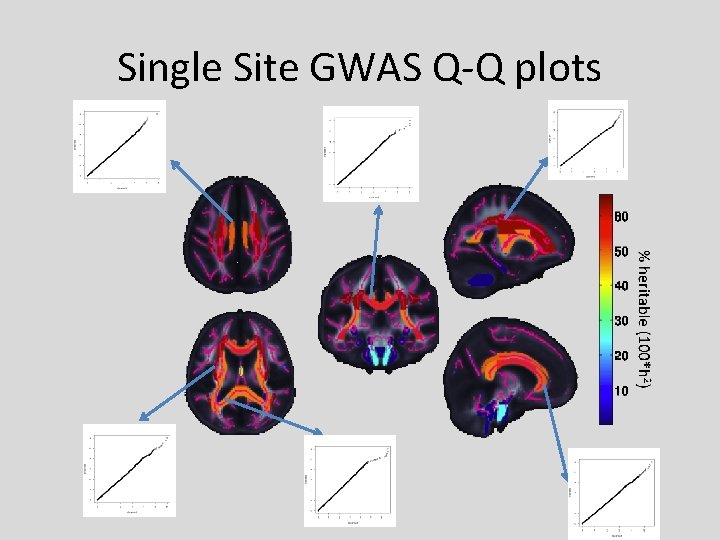
Single Site GWAS Q-Q plots

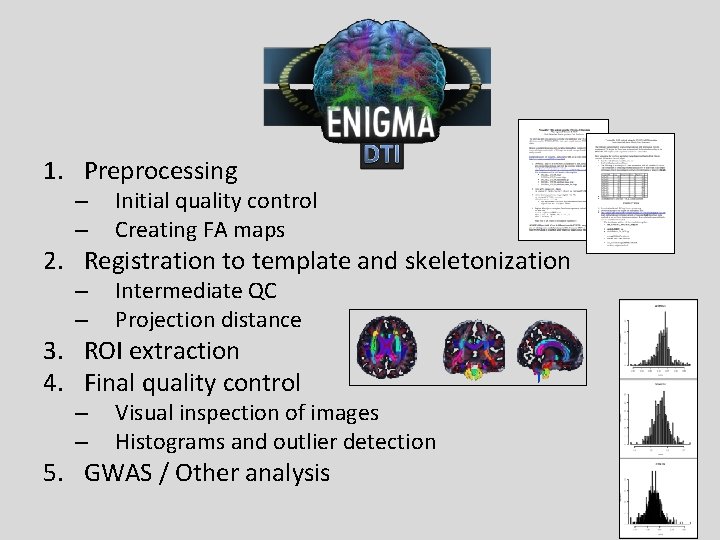
1. Preprocessing – – Initial quality control Creating FA maps 2. Registration to template and skeletonization – – Intermediate QC Projection distance 3. ROI extraction 4. Final quality control – – Visual inspection of images Histograms and outlier detection 5. GWAS / Other analysis

Acknowledgements ENIGMA DTI sites JOIN US Peter Kochunov 2#, Emma Sprooten 3, 4, René C. Mandl 5, Thomas E. Nichols 6, 7, Laura Almasy 8, John Blangero 8, Rachel M. Brouwer 4, Joanne E. Curran 8, Greig I. de Zubicaray 9, Ravi Duggirala 8, Peter T. Fox 10, L. Elliot Hong 2, Bennett A. Landman 11, Nicholas G. Martin 12, Katie L. Mc. Mahon 13, Sarah E. Medland 12, Braxton D. Mitchell 14, Rene L. Olvera 10, Charles P. Peterson 8, John M. Starr 15, Jessika E. Sussmann 4, Arthur W. Toga 1, Joanna M. Wardlaw 15, Margaret J. Wright 12, Hilleke E. Hulshoff Pol 5, Mark E. Bastin 4, 15, Andrew M. Mc. Intosh 4, Ian J. Deary 15, Paul M. Thompson 1*and David C. Glahn 3 US/Australia/European funding agencies Join ENIGMA http: //enigma. loni. ucla. edu/members/join/ Neda. Jahanshad@loni. ucla. edu