Multiple sequence alignment Why It is the most





- Slides: 45

Multiple sequence alignment Why? It is the most important means to assess relatedness n n n of a set of sequences Gain information about the structure/function of a query sequence (conservation patterns) Construct a phylogenetic tree Putting together a set of sequenced fragments (Fragment assembly) Recognise alternative splice sites Many bioinformatics methods depend on it (secondary/tertiary structure)

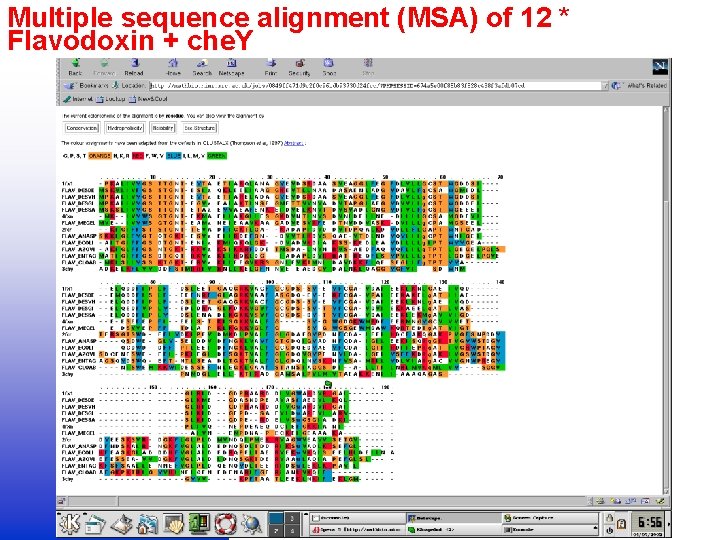
Multiple sequence alignment (MSA) of 12 * Flavodoxin + che. Y

Pairwise alignment n n n Now we know how to do it: How do we get a multiple alignment (three or more sequences)? Multiple alignment: much greater combinatorial explosion than with pairwise alignment…. .

Multi-dimensional dynamic programming (Murata et al. 1985)

Simultaneous Multiple alignment Multi-dimensional dynamic programming MSA (Lipman et al. , 1989, PNAS 86, 4412) n extremely slow and memory intensive n up to 8 -9 sequences of ~250 residues DCA (Stoye et al. , 1997, CABIOS 13, 625) n still very slow

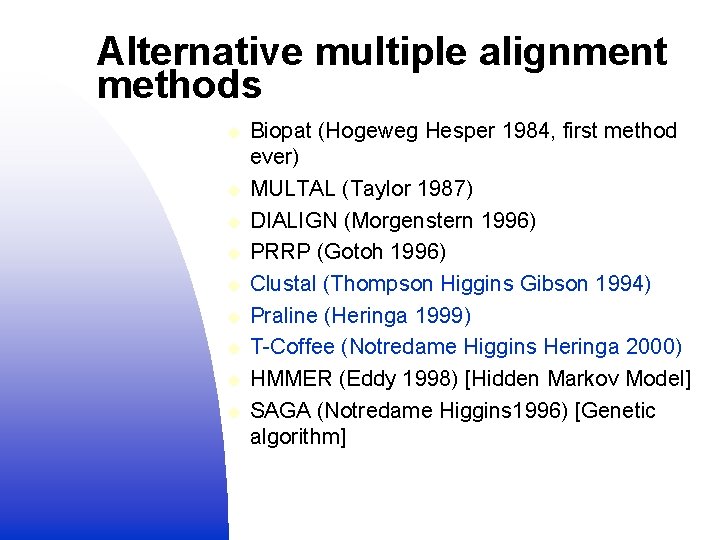
Alternative multiple alignment methods u u u u u Biopat (Hogeweg Hesper 1984, first method ever) MULTAL (Taylor 1987) DIALIGN (Morgenstern 1996) PRRP (Gotoh 1996) Clustal (Thompson Higgins Gibson 1994) Praline (Heringa 1999) T-Coffee (Notredame Higgins Heringa 2000) HMMER (Eddy 1998) [Hidden Markov Model] SAGA (Notredame Higgins 1996) [Genetic algorithm]

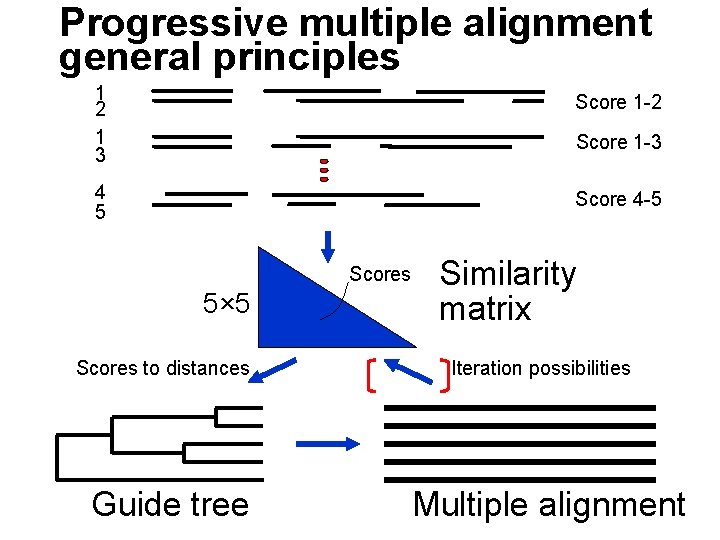
Progressive multiple alignment general principles 1 2 1 3 Score 1 -2 4 5 Score 4 -5 Score 1 -3 Scores 5× 5 Scores to distances Guide tree Similarity matrix Iteration possibilities Multiple alignment

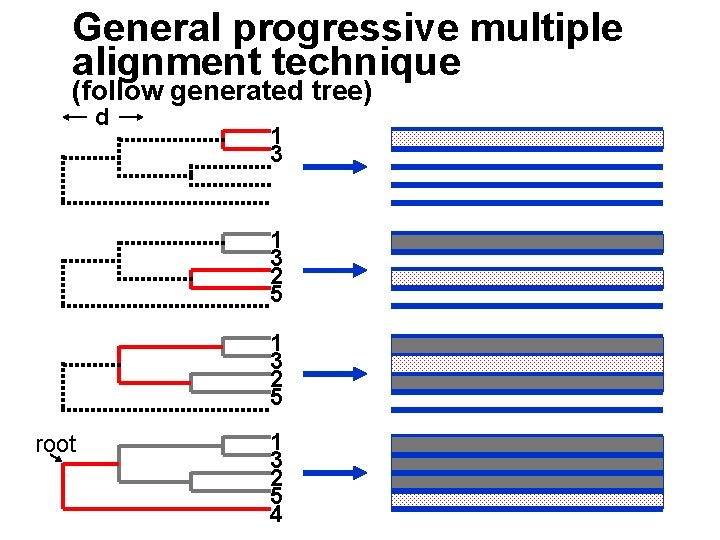
General progressive multiple alignment technique (follow generated tree) d 1 3 2 5 root 1 3 2 5 4

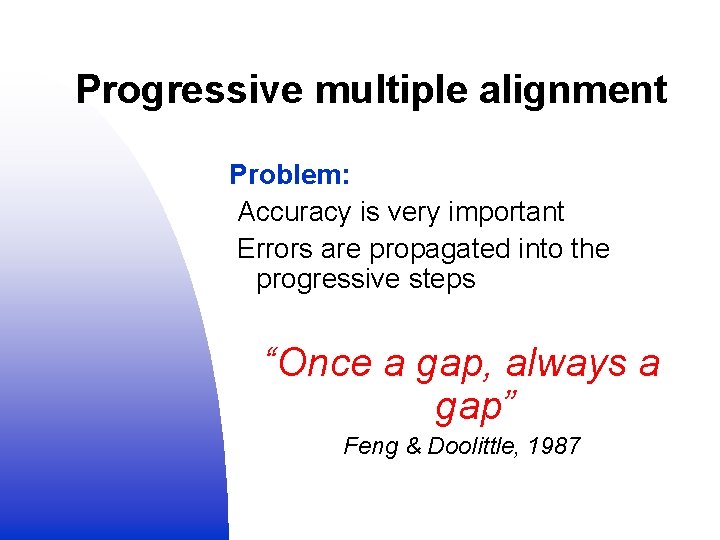
Progressive multiple alignment Problem: Accuracy is very important Errors are propagated into the progressive steps “Once a gap, always a gap” Feng & Doolittle, 1987

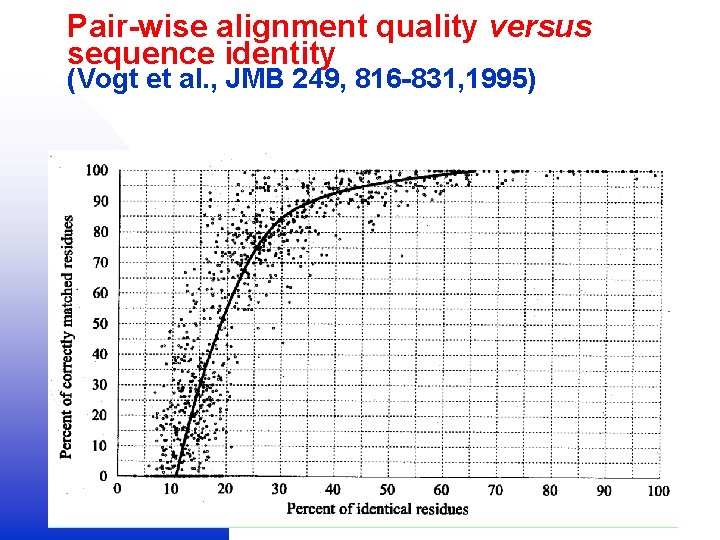
Pair-wise alignment quality versus sequence identity (Vogt et al. , JMB 249, 816 -831, 1995)

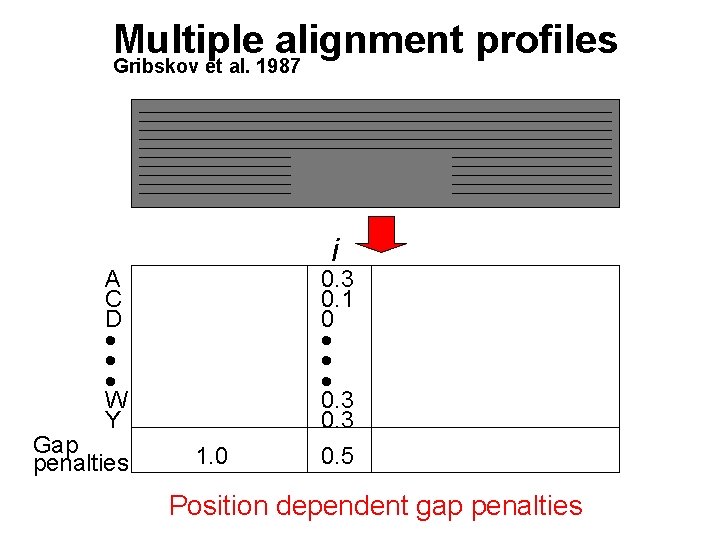
Multiple alignment profiles Gribskov et al. 1987 i A C D W Y Gap penalties 1. 0 0. 3 0. 1 0 0. 3 0. 5 Position dependent gap penalties

Profile-sequence alignment sequence profile ACD……VWY

Profile-profile alignment profile A C D. . Y profile ACD……VWY

Clustal, Clustal. W, Clustal. X n n n CLUSTAL W/X (Thompson et al. , 1994) uses Neighbour Joining (NJ) algorithm (Saitou and Nei, 1984), widely used in phylogenetic analysis, to construct guide tree. Sequence blocks are represented by profiles, in which the individual sequences are additionally weighted according to the branch lengths in the NJ tree. Further carefully crafted heuristics include: u u u n (i) local gap penalties (ii) automatic selection of the amino acid substitution matrix, (iii) automatic gap penalty adjustment (iv) mechanism to delay alignment of sequences that appear to be distant at the time they are considered. CLUSTAL (W/X) does not allow iteration (Hogeweg and Hesper, 1984; Corpet, 1988, Gotoh, 1996; Heringa, 1999, 2002)

Strategies for multiple sequence alignment n n Profile pre-processing Secondary structure-induced alignment Globalised local alignment Matrix extension Objective: try to avoid (early) errors

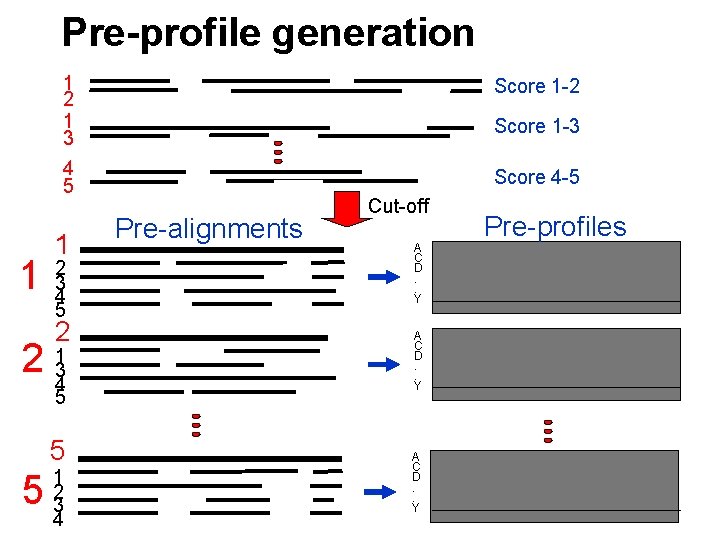
Pre-profile generation 1 1 2 1 3 Score 1 -2 4 5 Score 4 -5 1 2 3 4 5 2 2 134 5 5 5 1 2 3 4 Score 1 -3 Pre-alignments Cut-off A C D. . Y Pre-profiles

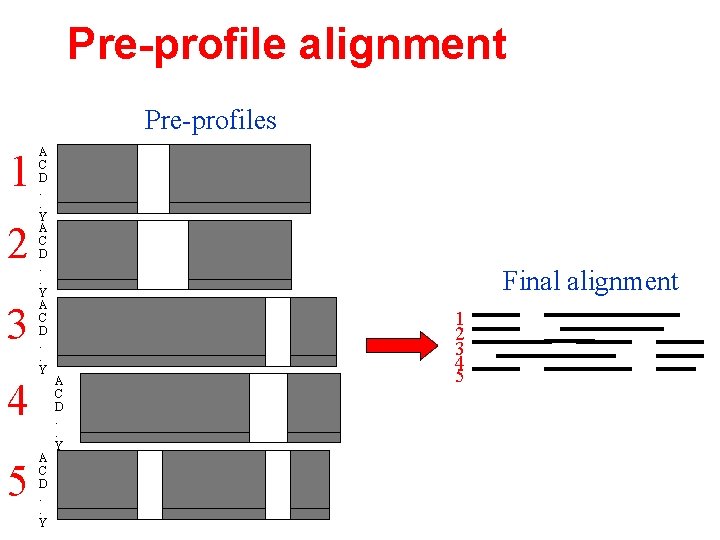
Pre-profile alignment Pre-profiles 1 2 3 4 5 A C D. . Y Final alignment A C D. . Y 1 2 3 4 5

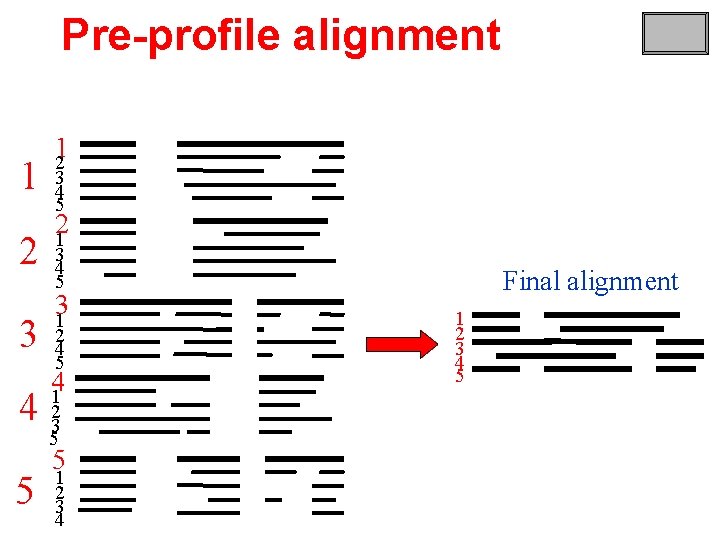
Pre-profile alignment 1 2 3 12 3 4 5 21 3 4 5 31 2 4 5 41 4 23 5 5 5 1 2 3 4 Final alignment 1 2 3 4 5

Strategies for multiple sequence alignment n n Profile pre-processing Secondary structure-induced alignment Globalised local alignment Matrix extension Objective: try to avoid (early) errors

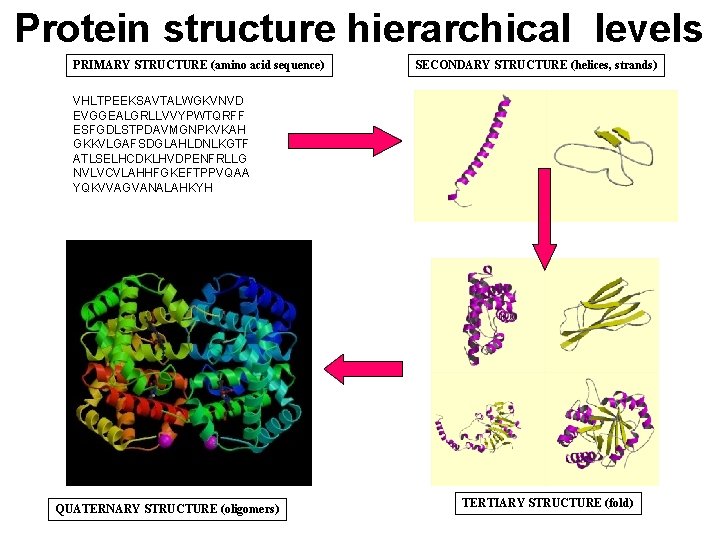
Protein structure hierarchical levels PRIMARY STRUCTURE (amino acid sequence) SECONDARY STRUCTURE (helices, strands) VHLTPEEKSAVTALWGKVNVD EVGGEALGRLLVVYPWTQRFF ESFGDLSTPDAVMGNPKVKAH GKKVLGAFSDGLAHLDNLKGTF ATLSELHCDKLHVDPENFRLLG NVLVCVLAHHFGKEFTPPVQAA YQKVVAGVANALAHKYH QUATERNARY STRUCTURE (oligomers) TERTIARY STRUCTURE (fold)

One of the Molecular Biology Dogma’s “Structure more conserved than sequence”

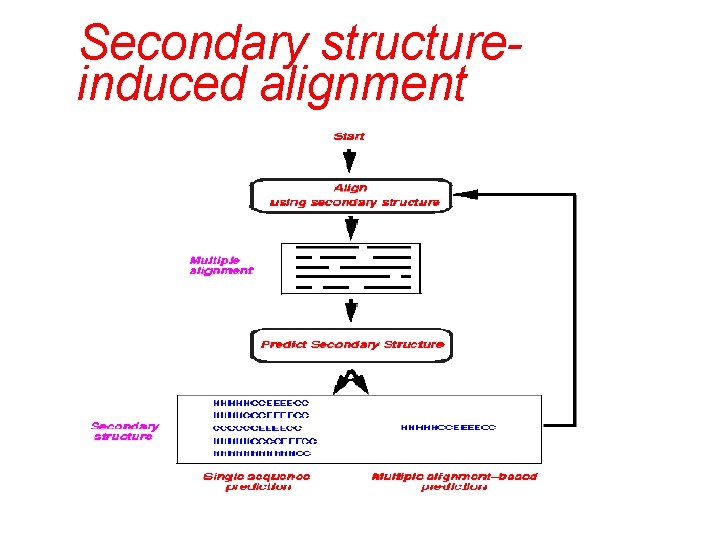
Secondary structureinduced alignment

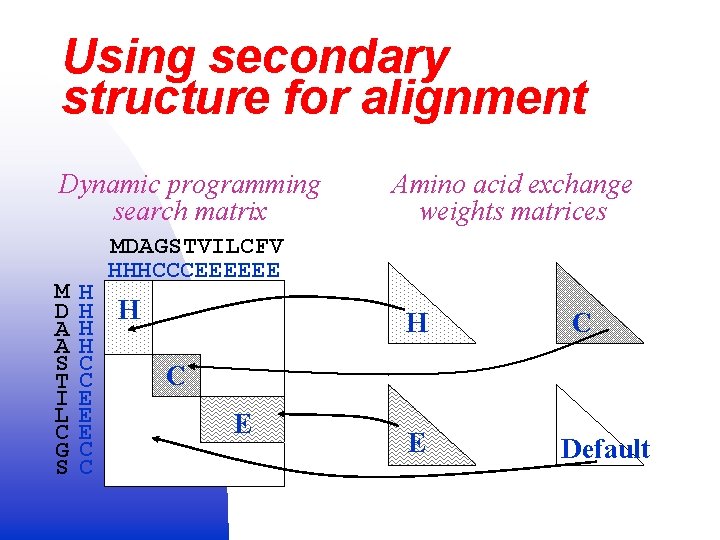
Using secondary structure for alignment Dynamic programming search matrix M D A A S T I L C G S Amino acid exchange weights matrices MDAGSTVILCFV HHHCCCEEEEEE H H H C C E E E C C H C C E E Default

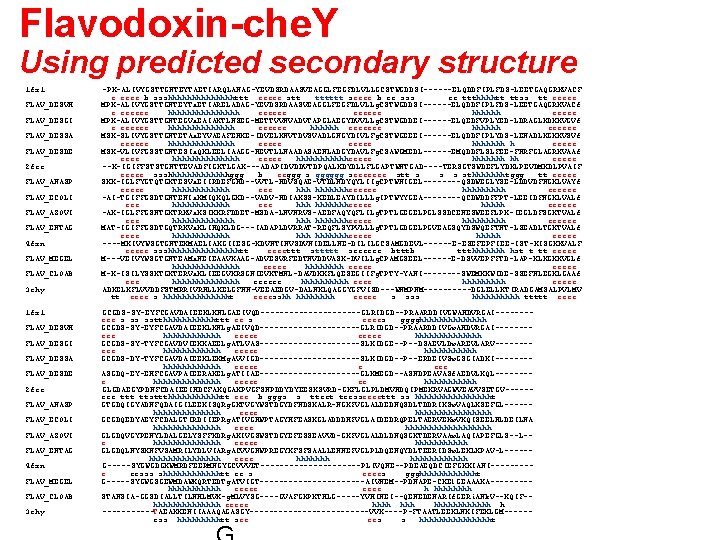
Flavodoxin-che. Y Using predicted secondary structure 1 fx 1 FLAV_DESVH FLAV_DESGI FLAV_DESSA FLAV_DESDE 2 fcr FLAV_ANASP FLAV_ECOLI FLAV_AZOVI FLAV_ENTAG 4 fxn FLAV_MEGEL FLAV_CLOAB 3 chy -PK-ALIVYGSTTGNTEYTAETIARQLANAG-YEVDSRDAASVEAGGLFEGFDLVLLGCSTWGDDSI------ELQDDFIPLFDS-LEETGAQGRKVACF e eeee b ssshhhhhhhttt eeeee stt tttttt seeee b ee sss ee ttthhhhtt ttss tt eeeee MPK-ALIVYGSTTGNTEYTa. ETIARELADAG-YEVDSRDAASVEAGGLFEGFDLVLLg. CSTWGDDSI------ELQDDFIPLFDS-LEETGAQGRKVACf e eeeeee hhhhhhhh eeeeee hhhhhh eeeee MPK-ALIVYGSTTGNTEGVa. EAIAKTLNSEG-METTVVNVADVTAPGLAEGYDVVLLg. CSTWGDDEI------ELQEDFVPLYED-LDRAGLKDKKVGVf e eeeeee hhhhhhh eeeeee MSK-SLIVYGSTTGNTETAa. EYVAEAFENKE-IDVELKNVTDVSVADLGNGYDIVLFg. CSTWGEEEI------ELQDDFIPLYDS-LENADLKGKKVSVf eeeeee hhhhhhh h eeeee MSK-VLIVFGSSTGNTESIa. QKLEELIAAGG-HEVTLLNAADASAENLADGYDAVLFg. CSAWGMEDL------EMQDDFLSLFEE-FNRFGLAGRKVAAf eeee hhhhhhh eeeee hhhhhheeeee hhhhhhh hh eeeee --K-IGIFFSTSTGNTTEVADFIGKTLGAK---ADAPIDVDDVTDPQALKDYDLLFLGAPTWNTGAD----TERSGTSWDEFLYDKLPEVDMKDLPVAIF eeeee ssshhhhhhhggg b eeggg s gggggg seeeeeee stt s sthhhhhhhtggg tt eeeee SKK-IGLFYGTQTGKTESVa. EIIRDEFGND--VVTL-HDVSQAE-VTDLNDYQYLIIg. CPTWNIGEL----QSDWEGLYSE-LDDVDFNGKLVAYf eeeee hhhhhhheeeeee hhhhh eeeeee -AI-TGIFFGSDTGNTENIa. KMIQKQLGKD--VADV-HDIAKSS-KEDLEAYDILLLg. IPTWYYGEA----QCDWDDFFPT-LEEIDFNGKLVALf eee hhhhhhheeeee hhhhh eeeeee -AK-IGLFFGSNTGKTRKVa. KSIKKRFDDET-MSDA-LNVNRVS-AEDFAQYQFLILg. TPTLGEGELPGLSSDCENESWEEFLPK-IEGLDFSGKTVALf eee hhhhhhheeeee hhhhh eeeeee MAT-IGIFFGSDTGQTRKVa. KLIHQKLDG---IADAPLDVRRAT-REQFLSYPVLLLg. TPTLGDGELPGVEAGSQYDSWQEFTNT-LSEADLTGKTVALf eeee hhhhhhheeeee hhhhh eeeee ----MKIVYWSGTGNTEKMAELIAKGIIESG-KDVNTINVSDVNIDELLNE-DILILGCSAMGDEVL------E-ESEFEPFIEE-IST-KISGKKVALF eeeee ssshhhhhhhhtt eeeettt sttttt seeeeee btttb ttthhhhhhh hst t tt eeeee M---VEIVYWSGTGNTEAMa. NEIEAAVKAAG-ADVESVRFEDTNVDDVASK-DVILLg. CPAMGSEEL------E-DSVVEPFFTD-LAP-KLKGKKVGLf hhhhhhh eeeee M-K-ISILYSSKTGKTERVa. KLIEEGVKRSGNIEVKTMNL-DAVDKKFLQESEGIIFg. TPTY-YANI----SWEMKKWIDE-SSEFNLEGKLGAAf eee hhhhhhh eeeeee hhhhh eeeee ADKELKFLVVDDFSTMRRIVRNLLKELGFNN-VEEAEDGV-DALNKLQAGGYGFVISD---WNMPNM-----DGLELLKTIRADGAMSALPVLMV tt eeee s hhhhhhht eeeesshh hhhh eeeee s sss hhhhh ttttt eeee GCGDS-SY-EYFCGAVDAIEEKLKNLGAEIVQD-----------GLRIDGD--PRAARDDIVGWAHDVRGAI-------eee s ss sstthhhhhhttt ee s eeees gggghhhhhhh GCGDS-SY-EYFCGAVDAIEEKLKNLg. AEIVQD-----------GLRIDGD--PRAARDDIVGw. AHDVRGAI-------eee hhhhhh eeeee hhhhhhh GCGDS-SY-TYFCGAVDVIEKKAEELg. ATLVAS-----------SLKIDGE--P--DSAEVLDw. AREVLARV-------eee hhhhhh eeeee hhhhhh GCGDS-DY-TYFCGAVDAIEEKLEKMg. AVVIGD-----------SLKIDGD--P--ERDEIVSw. GSGIADKI-------hhhhhh eeeee e eee ASGDQ-EY-EHFCGAVPAIEERAKELg. ATIIAE-----------GLKMEGD--ASNDPEAVASf. AEDVLKQL-------e hhhhhhh eeeee ee hhhhhh GLGDAEGYPDNFCDAIEEIHDCFAKQGAKPVGFSNPDDYDYEESKSVRD-GKFLGLPLDMVNDQIPMEKRVAGWVEAVVSETGV-----eee ttt ttsttthhhhhhtt eee b gggs s tteet teesseeeettt ss hhhhhhhht GTGDQIGYADNFQDAIGILEEKISQRg. GKTVGYWSTDGYDFNDSKALR-NGKFVGLALDEDNQSDLTDDRIKSw. VAQLKSEFGL-----hhhhhhh eeee hhhhhhhh GCGDQEDYAEYFCDALGTIRDIIEPRg. ATIVGHWPTAGYHFEASKGLADDDHFVGLAIDEDRQPELTAERVEKw. VKQISEELHLDEILNA hhhhhhh eeee hhhhhhhhh GLGDQVGYPENYLDALGELYSFFKDRg. AKIVGSWSTDGYEFESSEAVVD-GKFVGLALDLDNQSGKTDERVAAw. LAQIAPEFGLS--L-e hhhhhhh eeeee hhhhhh GLGDQLNYSKNFVSAMRILYDLVIARg. ACVVGNWPREGYKFSFSAALLENNEFVGLPLDQENQYDLTEERIDSw. LEKLKPAV-L-----hhhhhhhh eeee hhhhhhhhhhhh G-----SYGWGDGKWMRDFEERMNGYGCVVVET-----------PLIVQNE--PDEAEQDCIEFGKKIANI----e eesss shhhhhhtt ee s eeees ggghhhhhht G-----SYGWGSGEWMDAWKQRTEDTg. ATVIGT-----------AIVNEM--PDNAPE-CKEl. GEAAAKA----hhhhhh eeeee h hhhh STANSIA-GGSDIALLTILNHLMVK-g. MLVYSG----GVAFGKPKTHLG-----YVHINEI--QENEDENARIf. GERi. ANk. V--KQIF-hhhhhhh eeeee hhhhhh h ------TAEAKKENIIAAAQAGASGY-------------VVK----P-FTAATLEEKLNKIFEKLGM-----ess hhhhhtt see ees s hhhhhhhht

Strategies for multiple sequence alignment n Profile pre-processing Secondary structure-induced alignment n Globalised local alignment n Matrix extension n Objective: try to avoid (early) errors

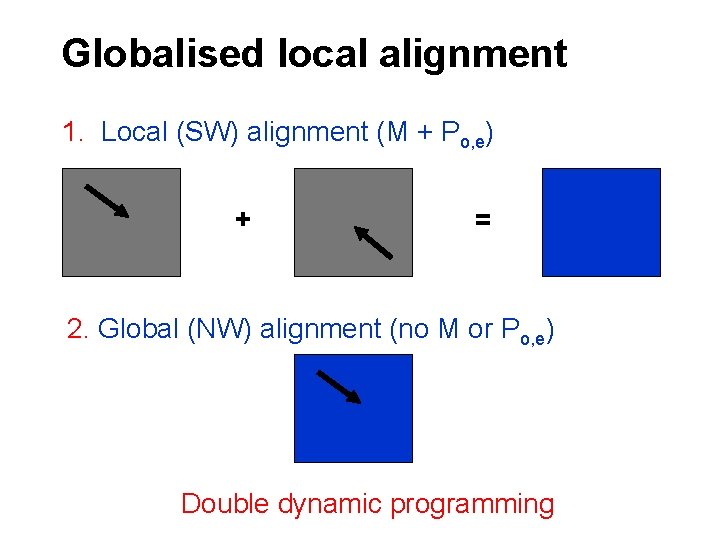
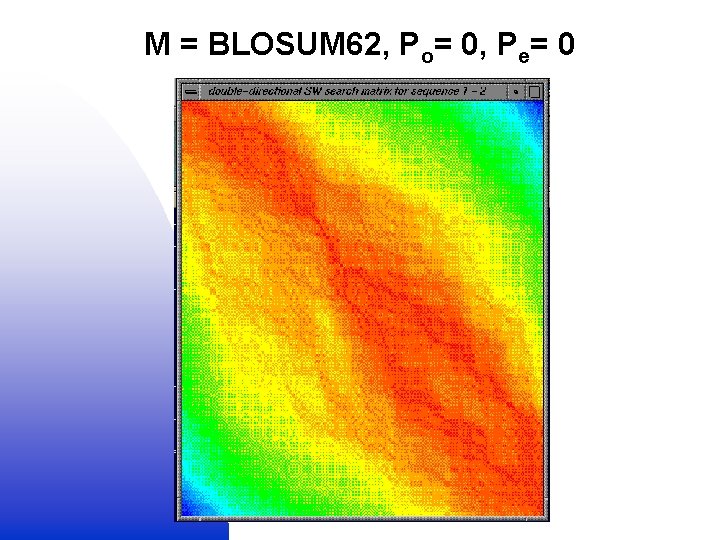
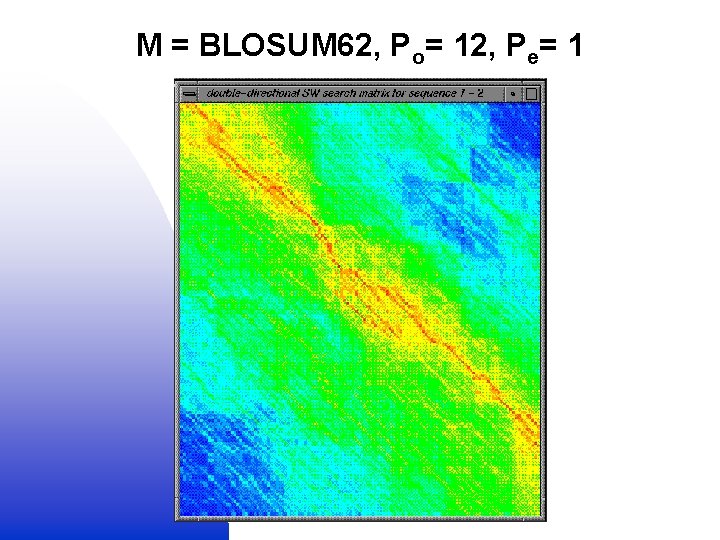
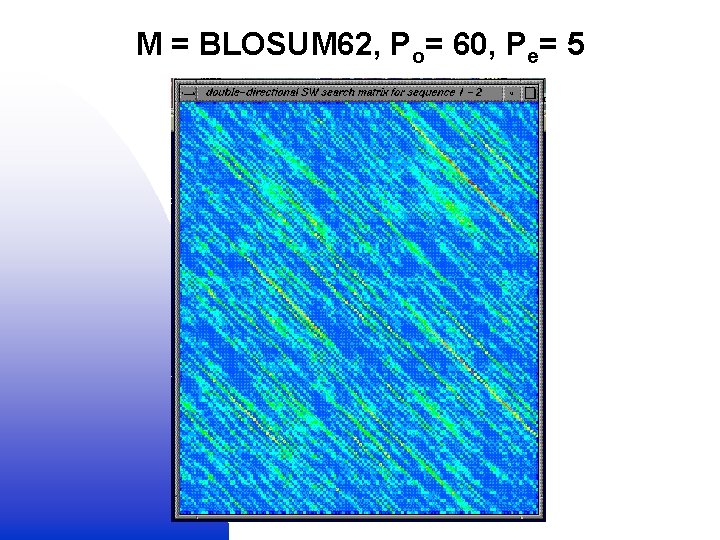
Globalised local alignment 1. Local (SW) alignment (M + Po, e) + = 2. Global (NW) alignment (no M or Po, e) Double dynamic programming

M = BLOSUM 62, Po= 0, Pe= 0

M = BLOSUM 62, Po= 12, Pe= 1

M = BLOSUM 62, Po= 60, Pe= 5

Strategies for multiple sequence alignment n Profile pre-processing Secondary structure-induced alignment Globalised local alignment n Matrix extension n n Objective: try to avoid (early) errors

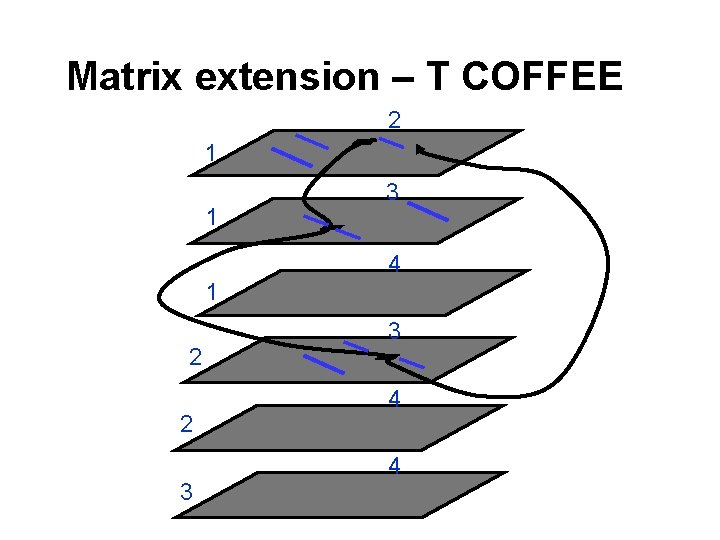
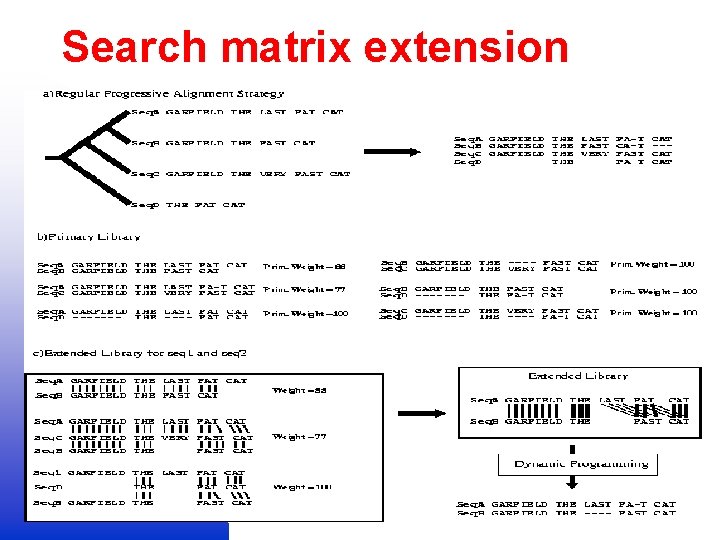
Matrix extension T-Coffee Tree-based Consistency Objective Function For alignm. Ent Evaluation Cedric Notredame Des Higgins Jaap Heringa J. Mol. Biol. , 302, 205 -217; 2000

Matrix extension – T COFFEE 2 1 1 3 4 1 2 2 3 3 4 4

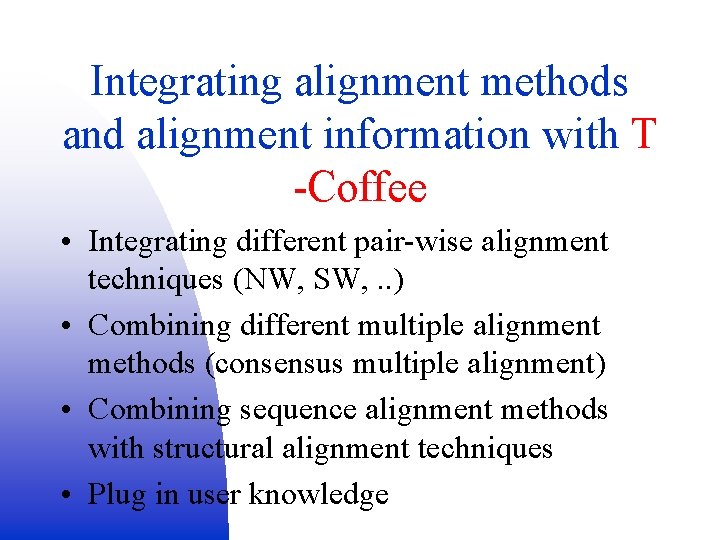
Integrating alignment methods and alignment information with T -Coffee • Integrating different pair-wise alignment techniques (NW, SW, . . ) • Combining different multiple alignment methods (consensus multiple alignment) • Combining sequence alignment methods with structural alignment techniques • Plug in user knowledge

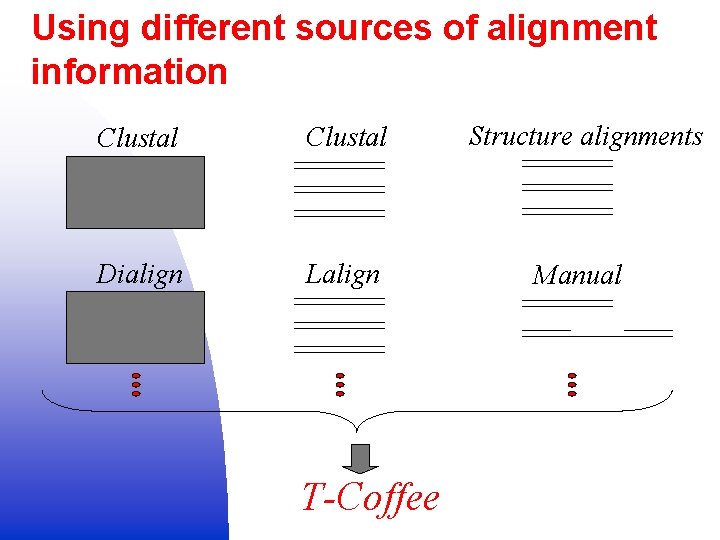
Using different sources of alignment information Clustal Structure alignments Dialign Lalign Manual T-Coffee

Search matrix extension

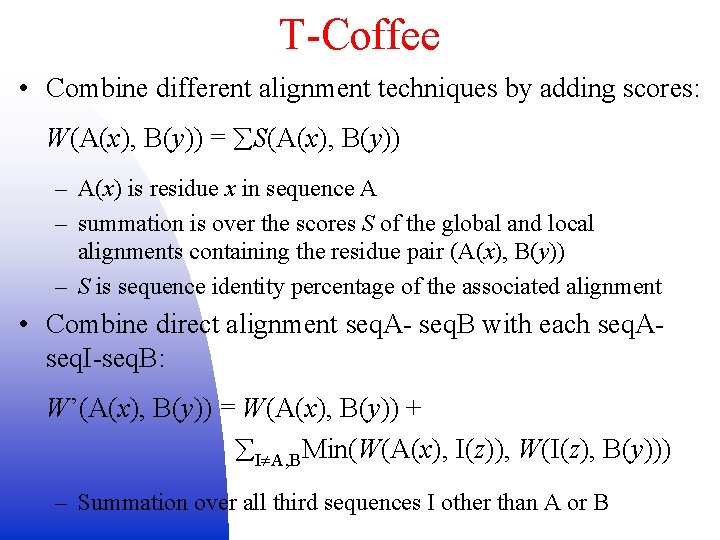
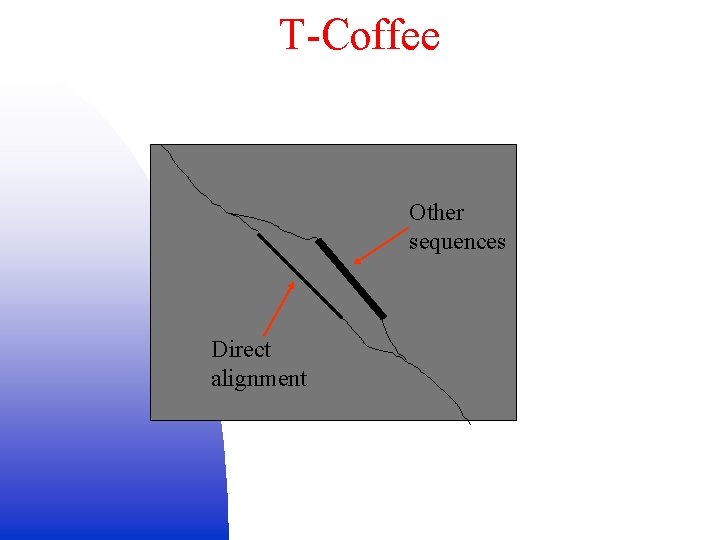
T-Coffee • Combine different alignment techniques by adding scores: W(A(x), B(y)) = S(A(x), B(y)) – A(x) is residue x in sequence A – summation is over the scores S of the global and local alignments containing the residue pair (A(x), B(y)) – S is sequence identity percentage of the associated alignment • Combine direct alignment seq. A- seq. B with each seq. Aseq. I-seq. B: W’(A(x), B(y)) = W(A(x), B(y)) + I A, BMin(W(A(x), I(z)), W(I(z), B(y))) – Summation over all third sequences I other than A or B

T-Coffee Other sequences Direct alignment

Search matrix extension

Evaluating multiple alignments n Conflicting standards of truth evolution u structure u function u n n n With orphan sequences no additional information Benchmarks depending on reference alignments Quality issue of available reference alignment databases Different ways to quantify agreement with reference alignment (sum-of-pairs, column score) “Charlie Chaplin” problem

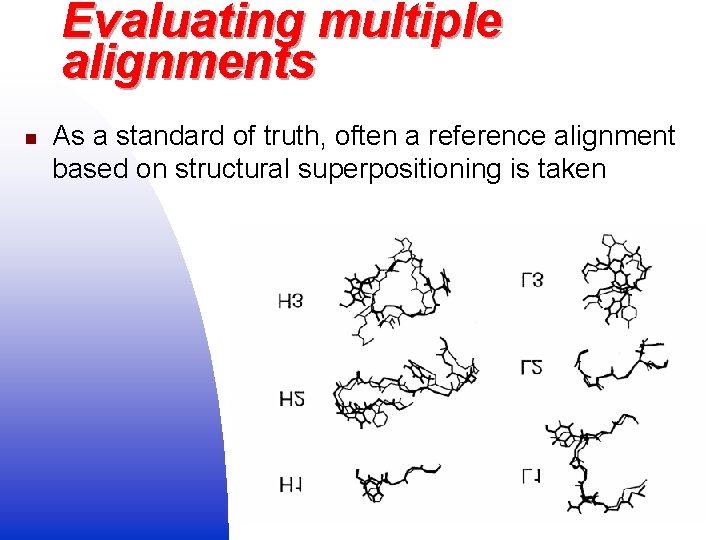
Evaluating multiple alignments n As a standard of truth, often a reference alignment based on structural superpositioning is taken

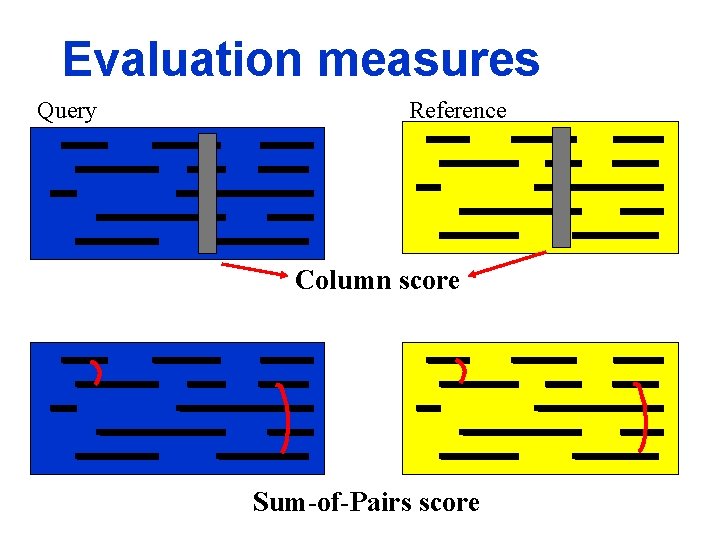
Evaluation measures Query Reference Column score Sum-of-Pairs score

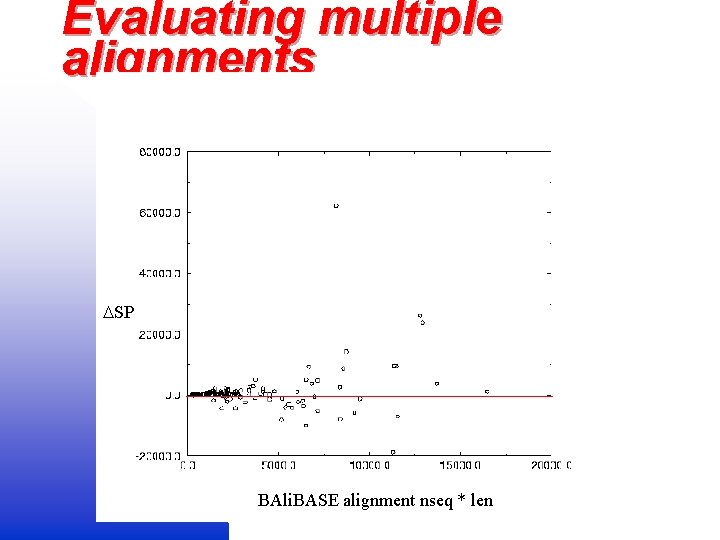
Evaluating multiple alignments SP BAli. BASE alignment nseq * len

Summary n Weighting schemes simulating simultaneous multiple alignment Profile pre-processing (global/local) u Matrix extension (well balanced scheme) u n Smoothing alignment signals u n Using additional information u n globalised local alignment secondary structure driven alignment Schemes strike balance between speed and sensitivity

References n n n Heringa, J. (1999) Two strategies for sequence comparison: profile-preprocessed and secondary structure-induced multiple alignment. Comp. Chem. 23, 341 -364. Notredame, C. , Higgins, D. G. , Heringa, J. (2000) TCoffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. , 302, 205 -217. Heringa, J. (2002) Local weighting schemes for protein multiple sequence alignment. Comput. Chem. , 26(5), 459 -477.

Where to find this…. http: //www. ibivu. cs. vu. nl/teaching