Multiple Sequence Alignment Dr Urmila KulkarniKale Bioinformatics Centre
Multiple Sequence Alignment Dr. Urmila Kulkarni-Kale Bioinformatics Centre University of Pune urmila@bioinfo. ernet. in Urmila. kulkarni. kale@gmail. com
Approaches: MSA • Dynamic programming • Progressive alignment: Clustal. W • Genetic algorithms: SAGA Jan 19, 2010 © UKK, Bioinformatics Centre, Uo. P 2
Progressive alignment approach • • • Align most related sequences Add on less related sequences to initial alignment Perform pairwise alignments of all sequences Use alignment scores to produce phylogenetic tree Align sequences sequentially, guided by the tree Gaps are added to an existing profile in progressive methods Jan 19, 2010 © UKK, Bioinformatics Centre, Uo. P 3
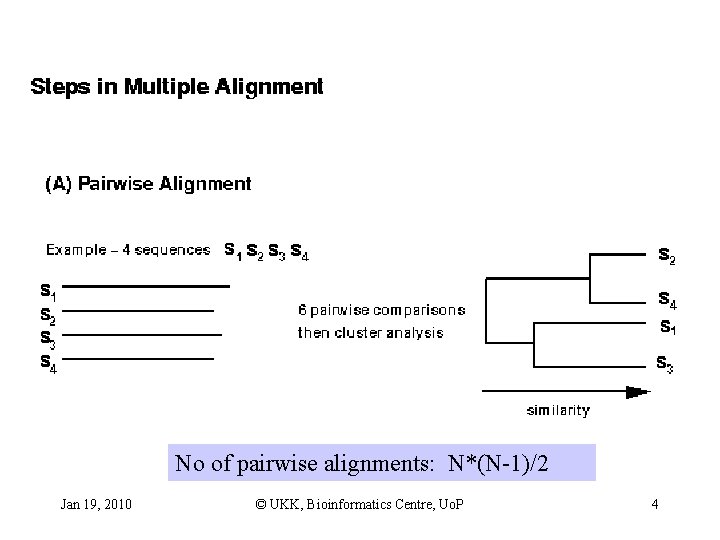
No of pairwise alignments: N*(N-1)/2 Jan 19, 2010 © UKK, Bioinformatics Centre, Uo. P 4
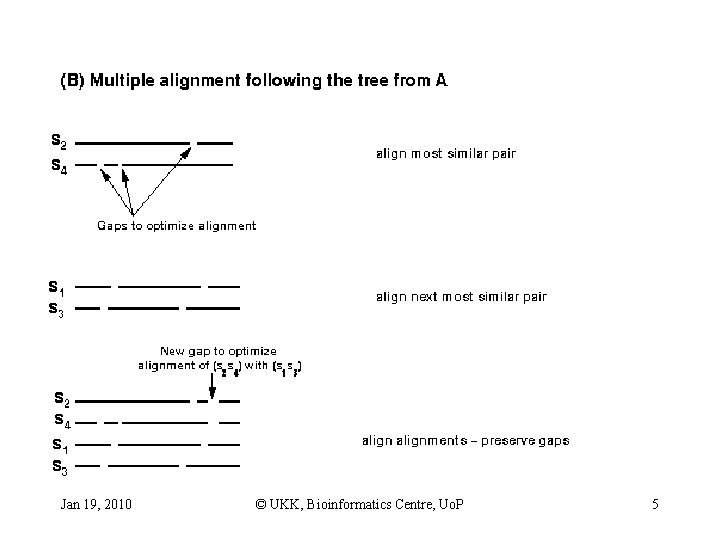
Jan 19, 2010 © UKK, Bioinformatics Centre, Uo. P 5
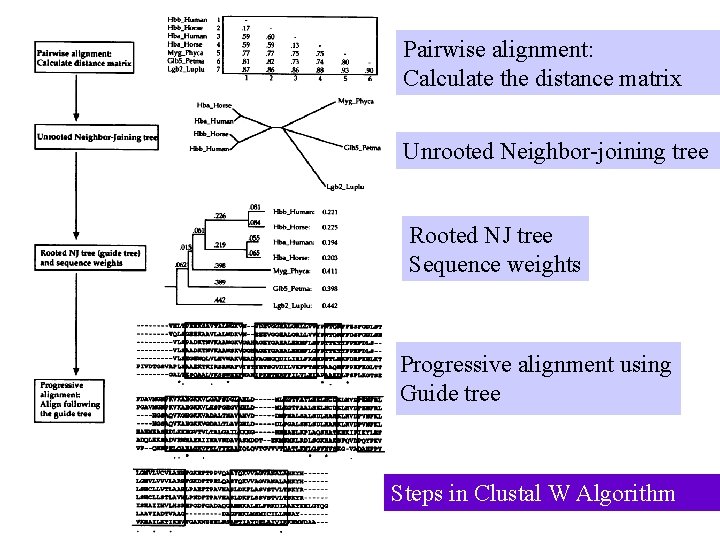
Pairwise alignment: Calculate the distance matrix Unrooted Neighbor-joining tree Rooted NJ tree Sequence weights Progressive alignment using Guide tree Jan 19, 2010 Steps in Clustal W Algorithm 6 © UKK, Bioinformatics Centre, Uo. P
Clustal W: weight • groups of related sequences receive lower weight • highly divergent sequences without any close relatives receive high weights Jan 19, 2010 © UKK, Bioinformatics Centre, Uo. P 7

Clustal. W: affine Gap penalty • GOP: gap opening penalty • GEP: gap extension penalty Heuristics in calculating gap penalty • Position specific penalty – gap at position? • yes lower GOP and GEP • no, but gap within 8 residues increase GOP – stretch of hydrophilic residues? • yes lower GOP • no use residue-specific gap propensities Once a gap, always a gap Jan 19, 2010 © UKK, Bioinformatics Centre, Uo. P 8
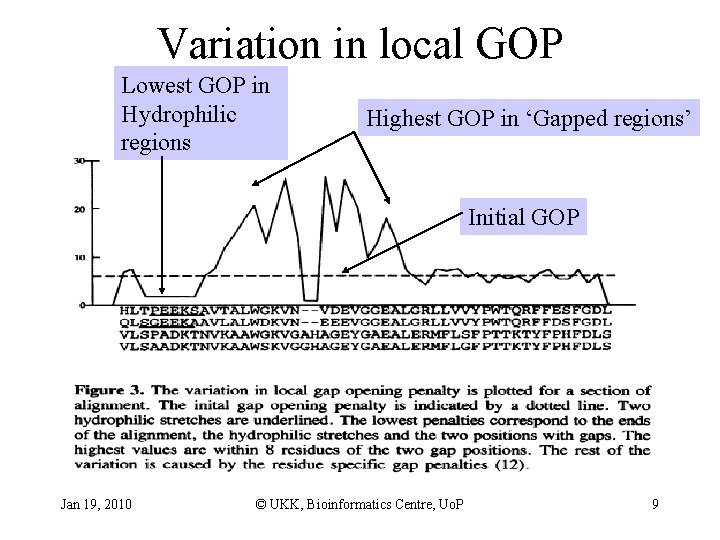
Variation in local GOP Lowest GOP in Hydrophilic regions Highest GOP in ‘Gapped regions’ Initial GOP Jan 19, 2010 © UKK, Bioinformatics Centre, Uo. P 9
Limitations of Progressive alignment approach • Greedy nature • Any errors in the initial alignment are carried through • More efficient for closely related sequences than for divergent sequences Jan 19, 2010 © UKK, Bioinformatics Centre, Uo. P 10
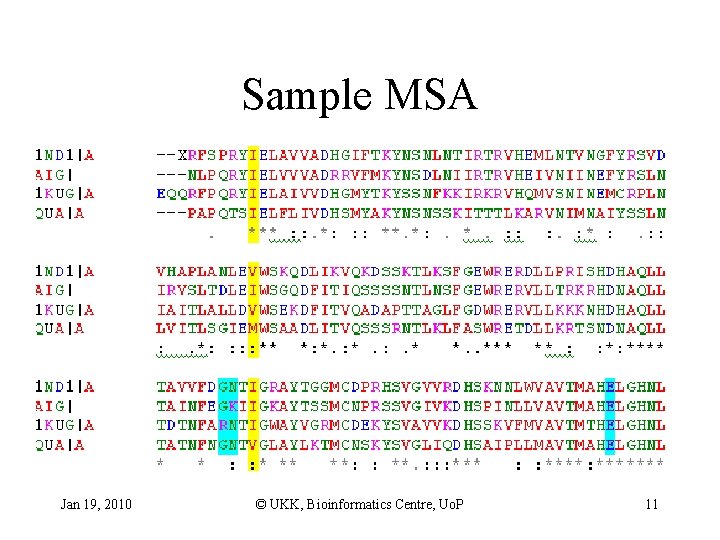
Sample MSA Jan 19, 2010 © UKK, Bioinformatics Centre, Uo. P 11
Applications of MSA • • • Detecting diagnostic patterns Phylogenetic analysis Primer design Prediction of protein secondary structure Finding novel relationships between genes Similar genes conserved across organisms – Same or similar function • Simultaneous alignment of similar genes yields: – regions subject to mutation – regions of conservation – mutations or rearrangements causing change in conformation or function Jan 19, 2010 © UKK, Bioinformatics Centre, Uo. P 12
- Slides: 12