Multiple Sequence Alignment Carlow IT Bioinformatics November 2006
Multiple Sequence Alignment Carlow IT Bioinformatics November 2006
MSA • A central technique in bioinformatics • along with: – homology searching – multiple sequence alignment – phylogenetic trees
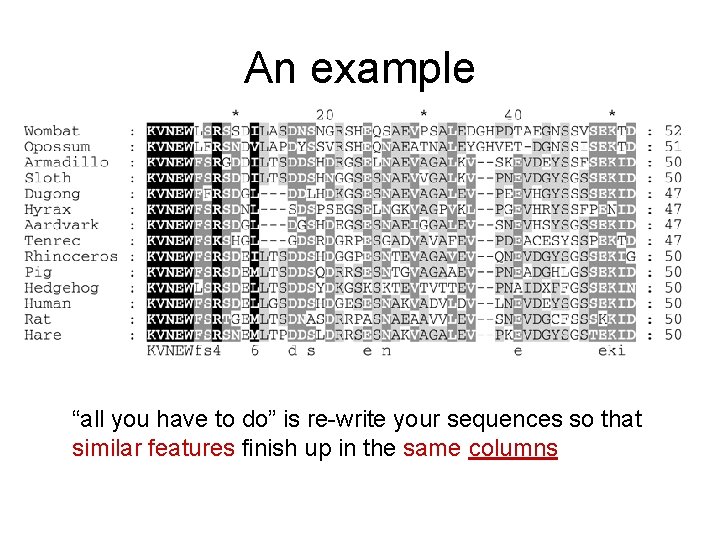
An example “all you have to do” is re-write your sequences so that similar features finish up in the same columns
Evolutionary relationship • “similar features” ideally means homologous – with a shared ancestor • clustal. W and T-coffee mimic the process of evolution – by weighting similar residues by how conserved they are in evolution • Important AAs don’t mutate • Less important AAs change easily, even randomly – by inserting judicious gaps

Criteria for alignment • Amino acids in the same column have – Structural similarity (used by threading progs) • Practical exercise inferring position of Bsu rec. A AAs – Evolutionary similarity – residues have a common ancestor – Functional similarity (active site, C-C bonds) may have to hand edit known functions – Sequence similarity • The first 3 (clear biological attributes) are, you hope, reflected by the last (an abstraction) which is what MSA programs use
Applications • Discover conserved patterns/motifs – A step to describing a protein domain – MSA can add a distant relative to your protein family • A step to define DNA regulatory elements. • Prediction of 2 nd Structure and helps 3 -D • A step to phylogenetic trees: to describe or show the process of evolution • PCR analysis/primer design – find most and least degenerate regions of your sequence
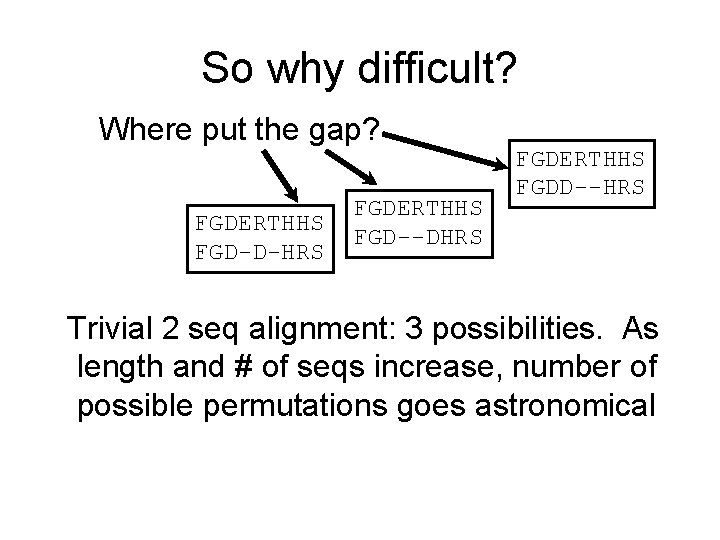
So why difficult? Where put the gap? FGDERTHHS FGD-D-HRS FGDERTHHS FGD--DHRS FGDERTHHS FGDD--HRS Trivial 2 seq alignment: 3 possibilities. As length and # of seqs increase, number of possible permutations goes astronomical

Some data • Cat ATGAAACGTCGGATCTAA • Dog ATGAATCGACCCATCTAA • Mus ATGGCGTGGCTTGGCATGTGA • Rat ATGGCATGTCGTGGCATGTAG Protocol step 1 • Align each pair of seqs C-D, C-M, C-R etc • Get a score for each alignment • And make a …
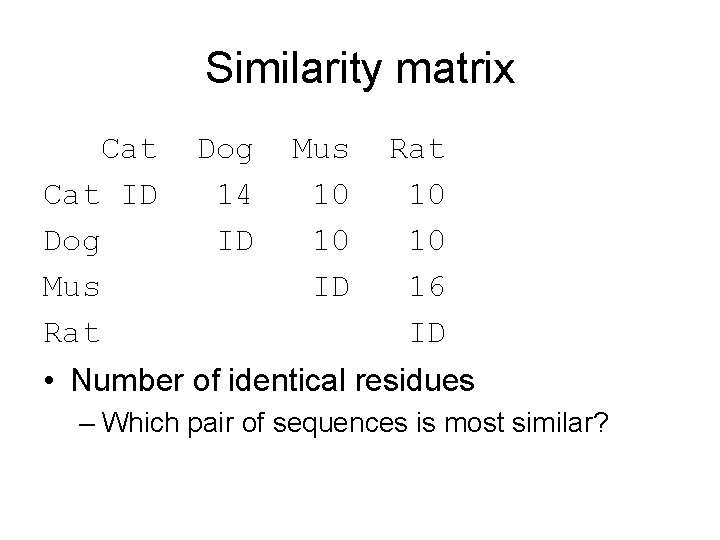
Similarity matrix Cat ID Dog Mus Rat Dog 14 ID Mus 10 10 ID Rat 10 10 16 ID • Number of identical residues – Which pair of sequences is most similar?

Progressive alignment • Align the two most similar sequences, inserting any gaps. • Mus/Rat: lock these sequences together (call it “RODent) • Return to similarity matrix to find next most similar seqs or sequence cluster • Dog/Cat: align and lock (call it CARnivore) – if next step requires a gap, then gap inserted in both carnivore sequences • Align next most …(now its iterative)
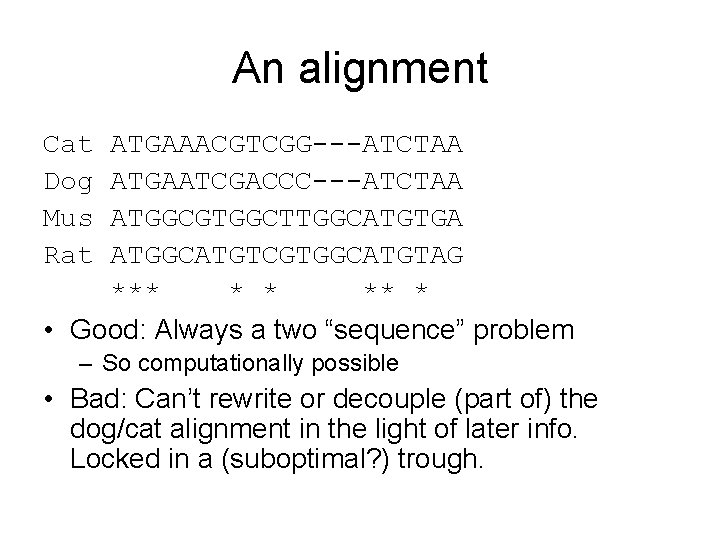
An alignment Cat Dog Mus Rat ATGAAACGTCGG---ATCTAA ATGAATCGACCC---ATCTAA ATGGCGTGGCTTGGCATGTGA ATGGCATGTCGTGGCATGTAG *** * * ** * • Good: Always a two “sequence” problem – So computationally possible • Bad: Can’t rewrite or decouple (part of) the dog/cat alignment in the light of later info. Locked in a (suboptimal? ) trough.
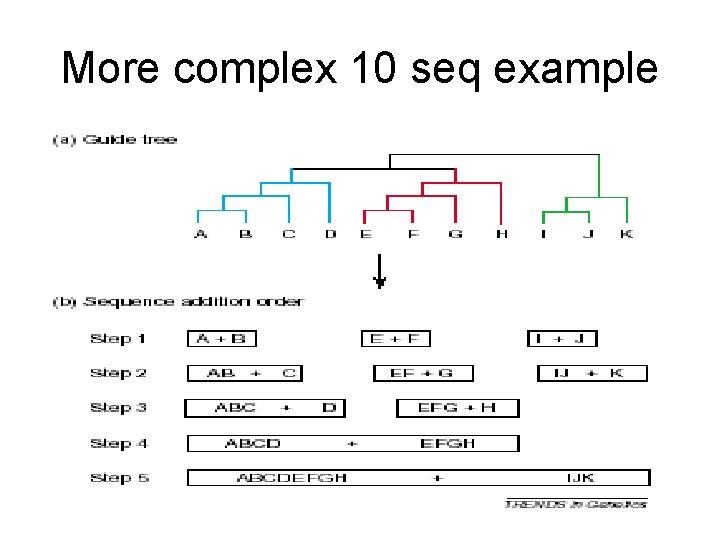
More complex 10 seq example

Choosing the right seqs • Use MSA to inform you! • Always use AA/protein if possible – can copygaps back to DNA later • • • Start with 6 -15 sequences Eliminate very different (<30% id) seqs Eliminate identical sequences Watch out for partial sequences …or sequences that need ++ gaps to align Check for repeats with dotlet, Lalign

Less is more • Large alignments – take ++ CPU and time – are hard to do well – are difficult to display – are difficult to use: in trees for example – may include marginal seqs that wreck whole alignment • So start small and add/eliminate seqs until you have a clear informative picture
Level of variation is important • Choose sequence family with best rate of evolution for your taxonomic group – Histones evolve very slow (compare kingdoms) – Transferrins are fast (compare classes, orders) • Closely related sequences may have identical protein (but variable DNA) • Distantly related sequences no DNA signal (“saturated”)
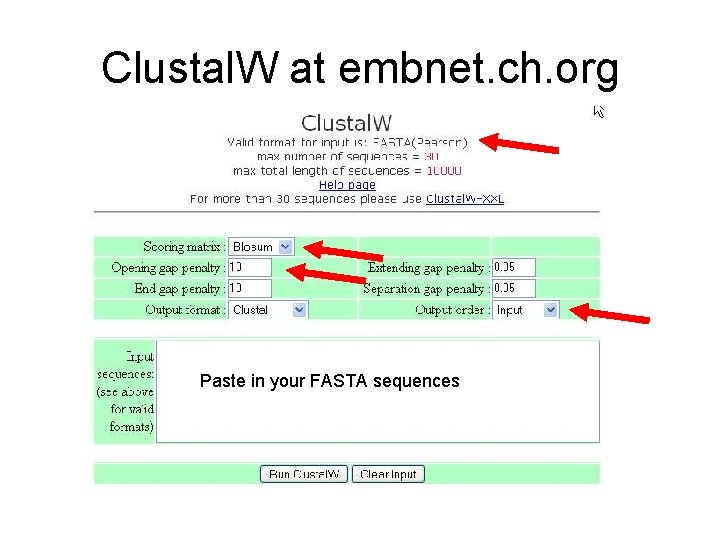
Clustal. W at embnet. ch. org Paste in your FASTA sequences
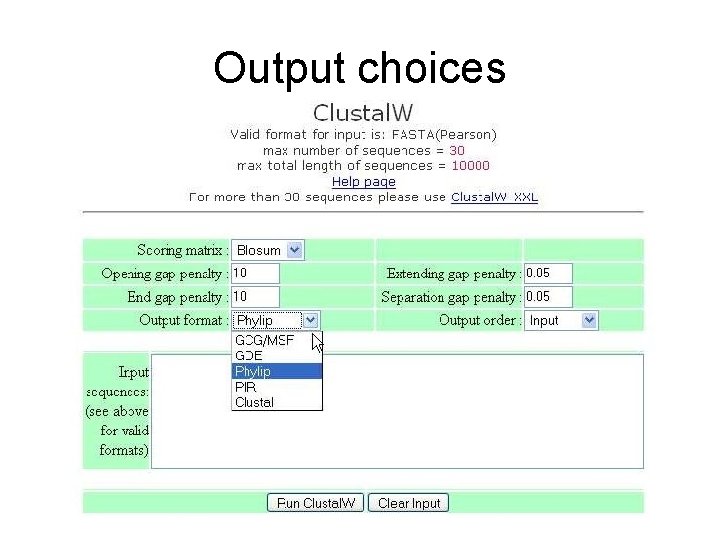
Output choices
Clustal. W at EBI Paste in your (FASTA) sequences
EBI: loads of options
T-coffee Minimal input parameters and STILL a better job than Clustal. W
Output EBI clustal. W Jalview alignment editor Pairwise distance etc Alignment Guidetree What you submitted
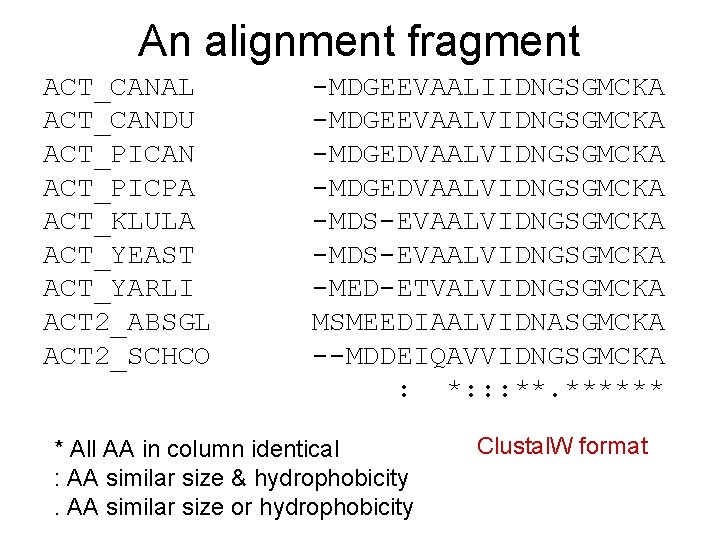
An alignment fragment ACT_CANAL ACT_CANDU ACT_PICAN ACT_PICPA ACT_KLULA ACT_YEAST ACT_YARLI ACT 2_ABSGL ACT 2_SCHCO -MDGEEVAALIIDNGSGMCKA -MDGEEVAALVIDNGSGMCKA -MDGEDVAALVIDNGSGMCKA -MDS-EVAALVIDNGSGMCKA -MED-ETVALVIDNGSGMCKA MSMEEDIAALVIDNASGMCKA --MDDEIQAVVIDNGSGMCKA : *: : : **. ****** * All AA in column identical : AA similar size & hydrophobicity. AA similar size or hydrophobicity Clustal. W format

The alignment, so what next? • • Look at it very closely Hand edit if necessary (probably) Eliminate problem sequences and redo? Use display option best for next step – Phylip format for trees
Parameter changes • Substit matrix PAM, Gonnet, Blosum – Clustalw chooses which matrix within family • PAM 30 for closely related pairs; PAM 120; PAM 250 for more distant – Difficult alignment: matrix change may help • Gap penalty (open and extend) have optimal values for each family: find which by trial and error. – Clustalw puts gaps (which are often external loops) near previous gaps (longer loop) • MSA does the grunt work. YOU do the fine tuning.
Guide tree • To figure which pairs of sequences to align first, a phylogenetic tree is calculated from pairwise distance matrix. – Stored in a DND (dendrogram) file • Never use this file to draw a tree • Clustalw can construct a tree from the multiple sequence alignment (better than pairwise)
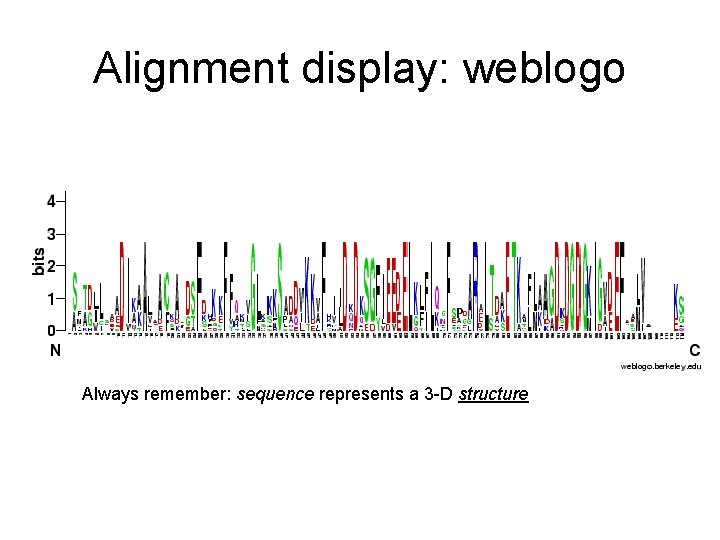
Alignment display: weblogo Always remember: sequence represents a 3 -D structure
Patterns to recognise (more reliable in MSA than in single seq) MSA improves 2 ndary structure (a-helix b-sheet) prediction by >6%) • Alternate hydrophobic residues – Surface b-sheet (zig-zag-zig-zag) • Runs of hydrophobic residues – Interior/buried b-sheet • Residues with 3. 5 AA spacing (amphipathic) – a-helix WNNWFNNFNNWNNNF • Gaps/indels – Probably surface not core

Conserved residues • W, F, Y large hydrophobic, internal/core – conserved WFY best signal for domains • G, P turns, can mark end of a-helix b-sheet • C conserved with reliable spacing speaks C-C disulphide bridges - defensins • H, S often catalytic sites in proteases (and other enzymes) • KRDE charged: ligand binding or salt-bridge • L very common AA but not conserved – except in Leucine zipper L 234567 L

Finish with an alignment: defensins 3 pairs of C residues: 3 disulphide bridges
- Slides: 29