Multiple Organ detection in CT Volumes Using Random





- Slides: 10

Multiple Organ detection in CT Volumes Using Random Forests - Week 5 Daniel Donenfeld

Work This week ● Test handcrafted features for classifying supervoxels Patch Features § Position and Histogram Features o Features at a Point § SIFT 3 D 1 § HOG 3 D 2 o 1. 2. Paul Scovanner, Saad Ali, and Mubarak Shah, A 3 -Dimensional SIFT Descriptor and its Application to Action Recognition, ACM MM 2007. Klaeser, A. , Marszalek, M. , & Schmid, C. (n. d. ). A Spatio-Temporal Descriptor Based on 3 D-Gradients. Procedings of the British Machine Vision Conference 2008.

Decision Tree ● Decision Trees Start at root, and go to child depending on decision rule o EX: if sunny then go to left node, otherwise go to right node o Leaves have labels for the data o To classify, start at root and apply decisions to data until reach the leaves o

Decision Tree

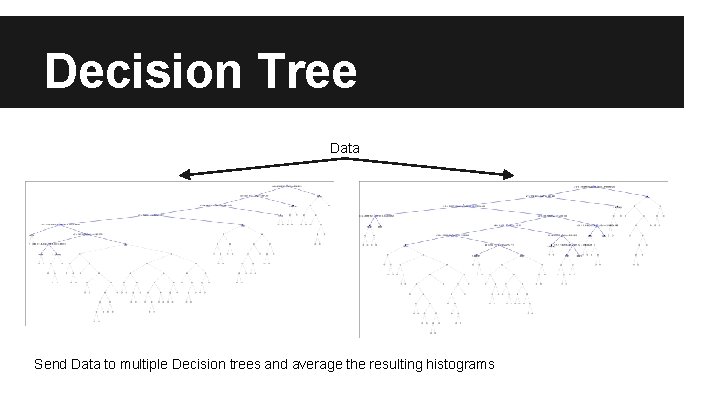
Random Forest ● Decision Trees - Prone to overfitting ● Random Forest - Create multiple decision trees from a subset of the data, using a random subset of features o o variance of each classifier reduces overfitting At each leaf node is a histogram of probabilities for each class

Decision Tree Data Send Data to multiple Decision trees and average the resulting histograms

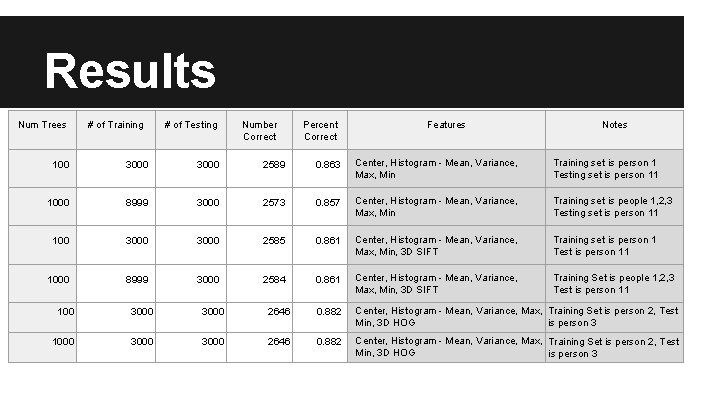
Results Num Trees # of Training # of Testing Number Correct Percent Correct Features Notes 100 3000 2589 0. 863 Center, Histogram - Mean, Variance, Max, Min Training set is person 1 Testing set is person 11 1000 8999 3000 2573 0. 857 Center, Histogram - Mean, Variance, Max, Min Training set is people 1, 2, 3 Testing set is person 11 100 3000 2585 0. 861 Center, Histogram - Mean, Variance, Max, Min, 3 D SIFT Training set is person 1 Test is person 11 1000 8999 3000 2584 0. 861 Center, Histogram - Mean, Variance, Max, Min, 3 D SIFT Training Set is people 1, 2, 3 Test is person 11 100 3000 2646 0. 882 Center, Histogram - Mean, Variance, Max, Training Set is person 2, Test Min, 3 D HOG is person 3 1000 3000 2646 0. 882 Center, Histogram - Mean, Variance, Max, Training Set is person 2, Test Min, 3 D HOG is person 3

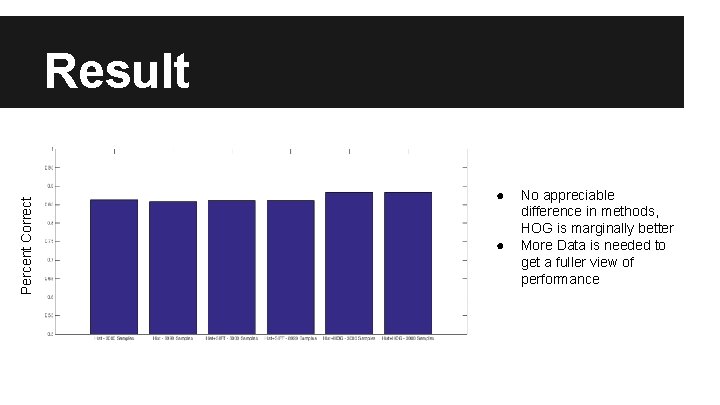
Percent Correct Result ● ● No appreciable difference in methods, HOG is marginally better More Data is needed to get a fuller view of performance

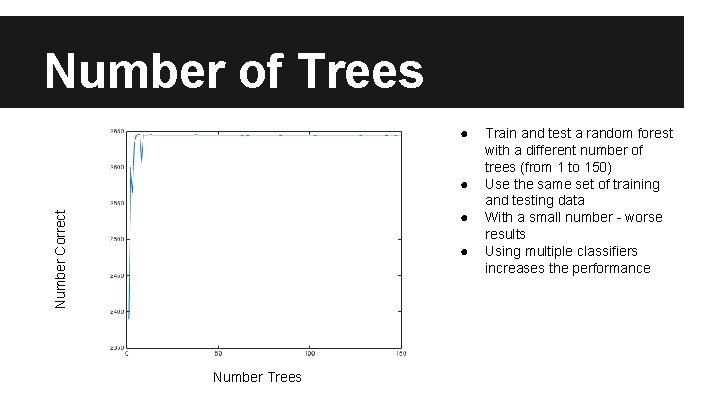
Number of Trees ● ● Number Correct ● ● Number Trees Train and test a random forest with a different number of trees (from 1 to 150) Use the same set of training and testing data With a small number - worse results Using multiple classifiers increases the performance

Next Week Plan ● Still running code on complete dataset o Supervoxel code takes ~8 hours/patient ● Continue testing 3 DSIFT and 3 DHOG ● Test other 3 D image features o 3 D gabor filters for texture information ● Improve classification using distance priors