Multiple Global Sequence Alignment and Phylogenetic trees Inge
Multiple Global Sequence Alignment and Phylogenetic trees Inge Jonassen and Ingvar Eidhammer et al. Protein Bioinformatics Chapter 4 1

Definition • A global alignment of a set of sequences is obtained by – inserting into each sequence gap characters ‘ ’ • so that – the resulting sequences are of the same length • and so that – no “column” has only gap characters Eidhammer et al. Protein Bioinformatics Chapter 4 2
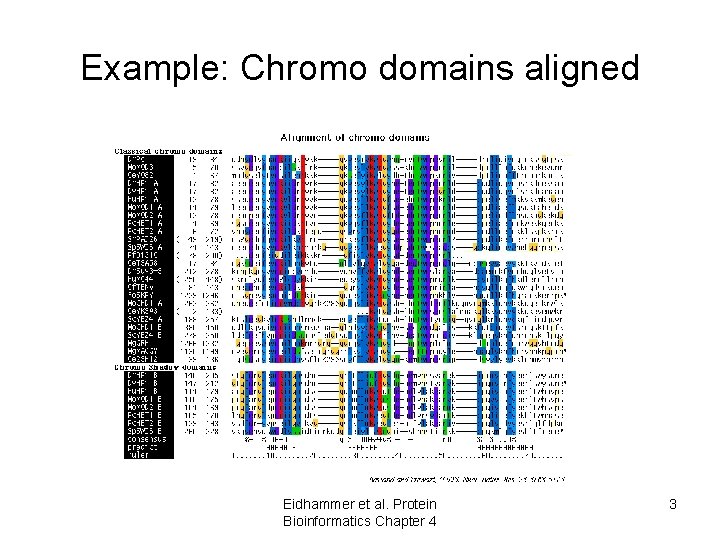
Example: Chromo domains aligned Eidhammer et al. Protein Bioinformatics Chapter 4 3
Use of alignments • High sequence similarity usually means significant structural and/or functional similarity. The reverse does not need to be true • Homolog proteins (common ancestor) can vary significantly in large parts of the sequences, but still retain common 2 D-patterns, 3 Dpatterns or common active site or binding site. • Comparison of several sequences in a family can reveal what is common for the family (From Lesk: Two homologous sequences whisper, . . A full multiple alignment shouts out load). Something common for several sequences can be significant when regarding all of the sequences, but need not if regarding only two. • Multiple alignment can be used to derive evolutionary history. Eidhammer et al. Protein Bioinformatics Chapter 4 4
Use of alignments • Predict features of aligned objects – conserved positions • structurally/functionally important Eidhammer et al. Protein Bioinformatics Chapter 4 5
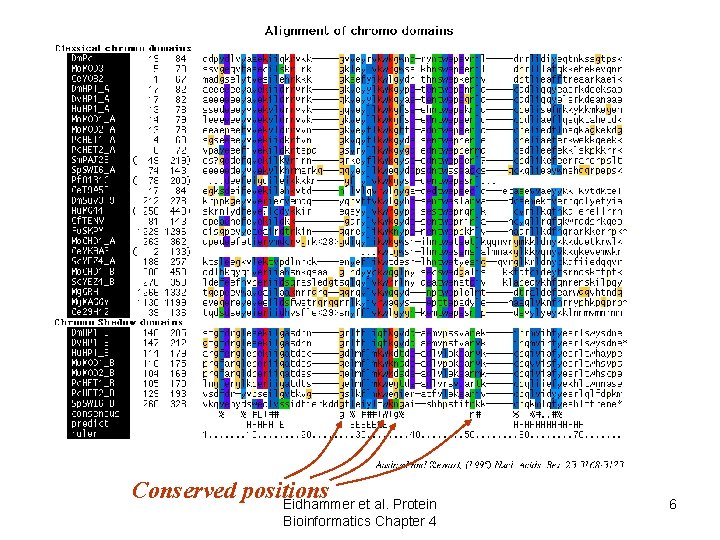
Conserved positions Eidhammer et al. Protein Bioinformatics Chapter 4 6
Use of alignments • Predict features of aligned objects – conserved positions • structurally/functionally important – patterns of hydrophobicity/hydrophilicity • secondary structure elements Eidhammer et al. Protein Bioinformatics Chapter 4 7
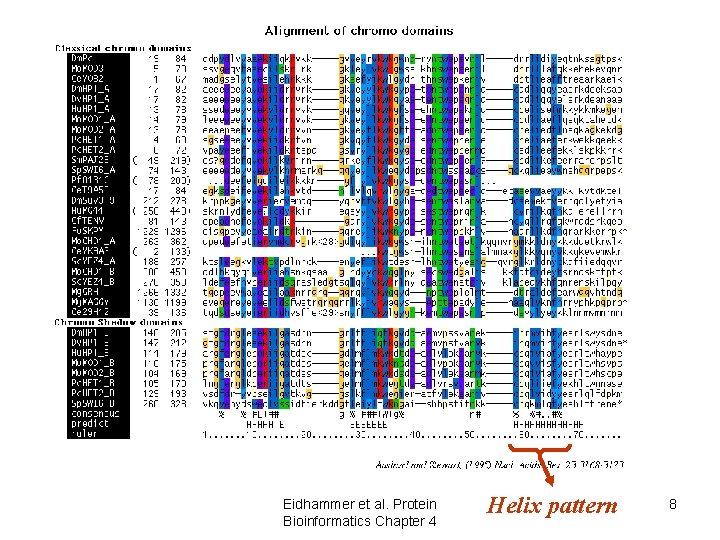
Eidhammer et al. Protein Bioinformatics Chapter 4 Helix pattern 8
Use of alignments • Predict features of aligned objects – conserved positions • structurally/functionally important – patterns of hydrophobicity/hydrophilicity • secondary structure elements – “gappy” regions • loops/variable regions Eidhammer et al. Protein Bioinformatics Chapter 4 9
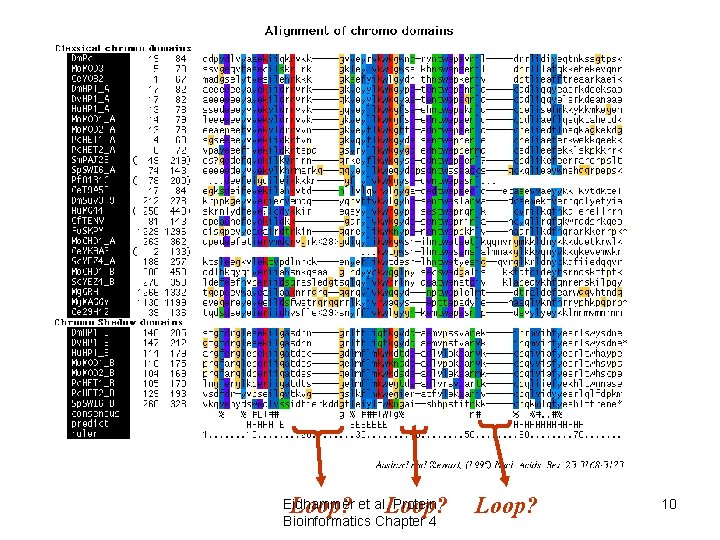
Loop? Eidhammer et al. Protein Bioinformatics Chapter 4 Loop? 10
Use of alignments • Predict features of aligned objects – conserved positions • structurally/functionally important – patterns of hydrophobicity/hydrophilicity • secondary structure elements – “gappy” regions • loops/variable regions – covariation • structural proximity Eidhammer et al. Protein Bioinformatics Chapter 4 11
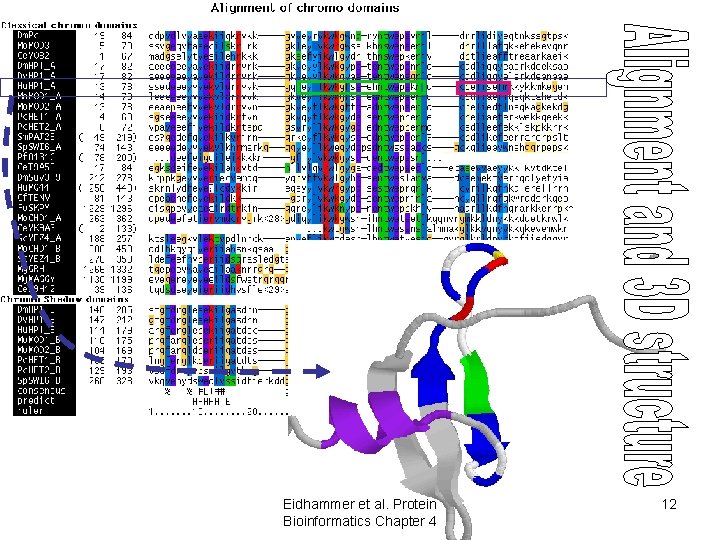
Eidhammer et al. Protein Bioinformatics Chapter 4 12
Use of Alignments - make patterns/profiles • Can make a profile or a pattern that can be used to match against a sequence database and identify new family members • Profiles/patterns can be used to predict family membership of new sequences • Databases of profiles/patterns – PROSITE – PFAM – PRINTS –. . . Eidhammer et al. Protein Bioinformatics Chapter 4 13
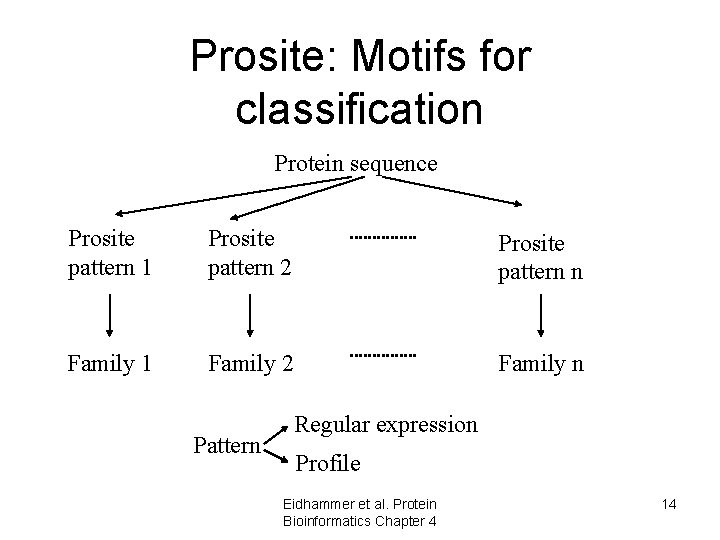
Prosite: Motifs for classification Protein sequence Prosite pattern 1 Prosite pattern 2 Prosite pattern n Family 1 Family 2 Family n Pattern Regular expression Profile Eidhammer et al. Protein Bioinformatics Chapter 4 14
![Pattern from alignment [FYL]-x-[LIVMC]-[KR]-W-x-[GDNR]-[FYWLE]-x(5, 6)-[ST]-W-[ES]-[PSTDN]-x(3)-[LIVMC] Eidhammer et al. Protein Bioinformatics Chapter 4 15 Pattern from alignment [FYL]-x-[LIVMC]-[KR]-W-x-[GDNR]-[FYWLE]-x(5, 6)-[ST]-W-[ES]-[PSTDN]-x(3)-[LIVMC] Eidhammer et al. Protein Bioinformatics Chapter 4 15](http://slidetodoc.com/presentation_image/db6195ed1136096e04e057e7250a927d/image-15.jpg)
Pattern from alignment [FYL]-x-[LIVMC]-[KR]-W-x-[GDNR]-[FYWLE]-x(5, 6)-[ST]-W-[ES]-[PSTDN]-x(3)-[LIVMC] Eidhammer et al. Protein Bioinformatics Chapter 4 15
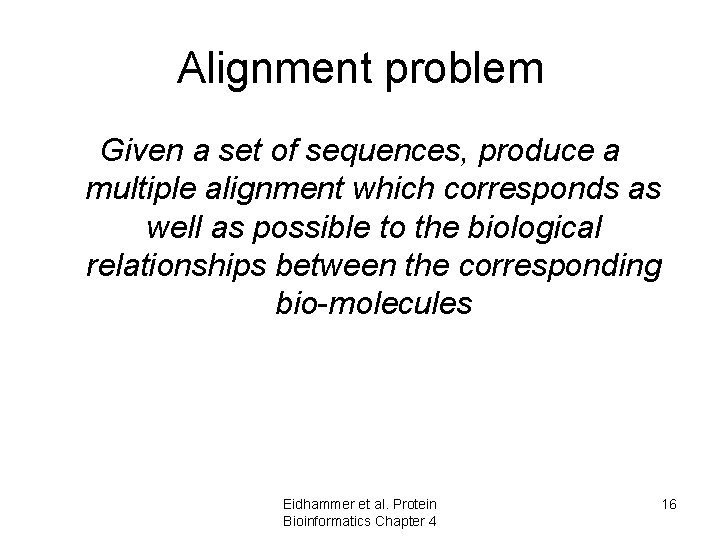
Alignment problem Given a set of sequences, produce a multiple alignment which corresponds as well as possible to the biological relationships between the corresponding bio-molecules Eidhammer et al. Protein Bioinformatics Chapter 4 16

For homologous proteins • Two residues should be aligned (on top of each other) – if they are homologous (evolved from the same residue in a common ancestor protein) – if they are structurally equivalent Eidhammer et al. Protein Bioinformatics Chapter 4 17
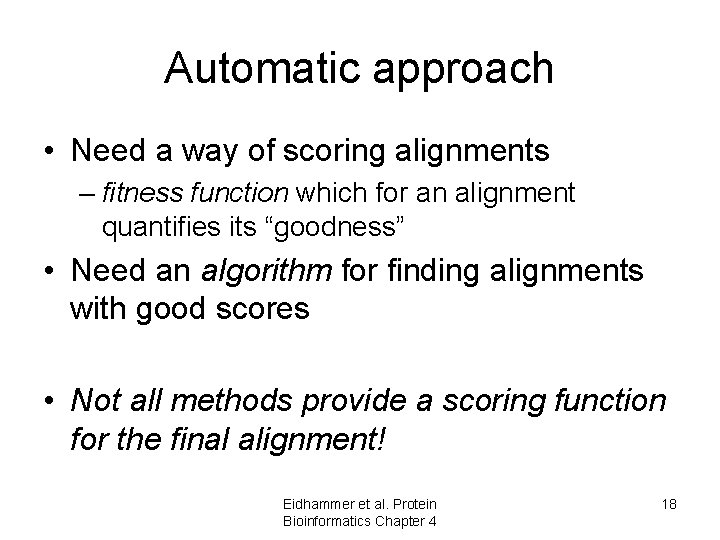
Automatic approach • Need a way of scoring alignments – fitness function which for an alignment quantifies its “goodness” • Need an algorithm for finding alignments with good scores • Not all methods provide a scoring function for the final alignment! Eidhammer et al. Protein Bioinformatics Chapter 4 18
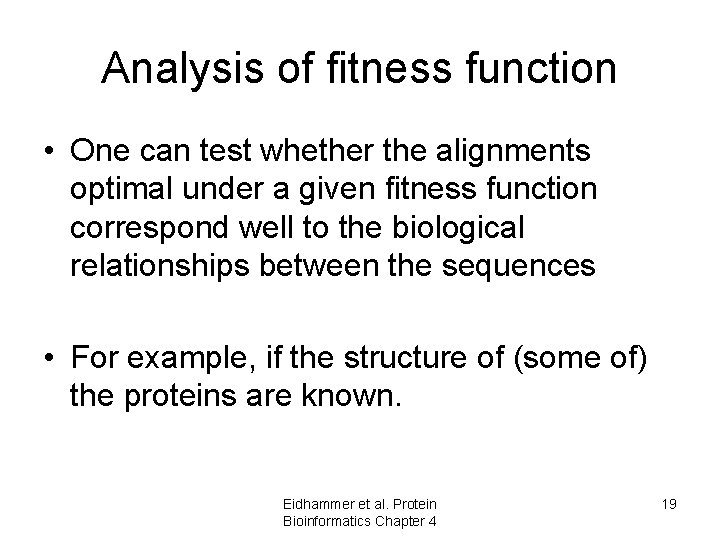
Analysis of fitness function • One can test whether the alignments optimal under a given fitness function correspond well to the biological relationships between the sequences • For example, if the structure of (some of) the proteins are known. Eidhammer et al. Protein Bioinformatics Chapter 4 19
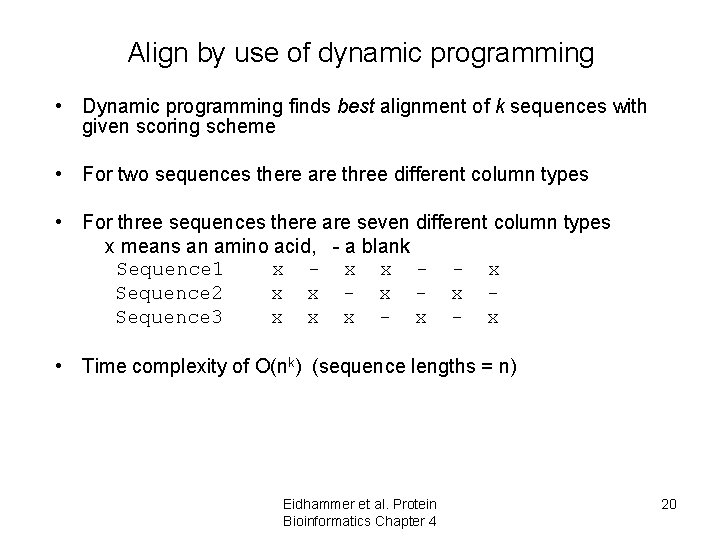
Align by use of dynamic programming • Dynamic programming finds best alignment of k sequences with given scoring scheme • For two sequences there are three different column types • For three sequences there are seven different column types x means an amino acid, - a blank Sequence 1 x - x x - - x Sequence 2 x x - x Sequence 3 x x x - x • Time complexity of O(nk) (sequence lengths = n) Eidhammer et al. Protein Bioinformatics Chapter 4 20

Eidhammer et al. Protein Bioinformatics Chapter 4 21
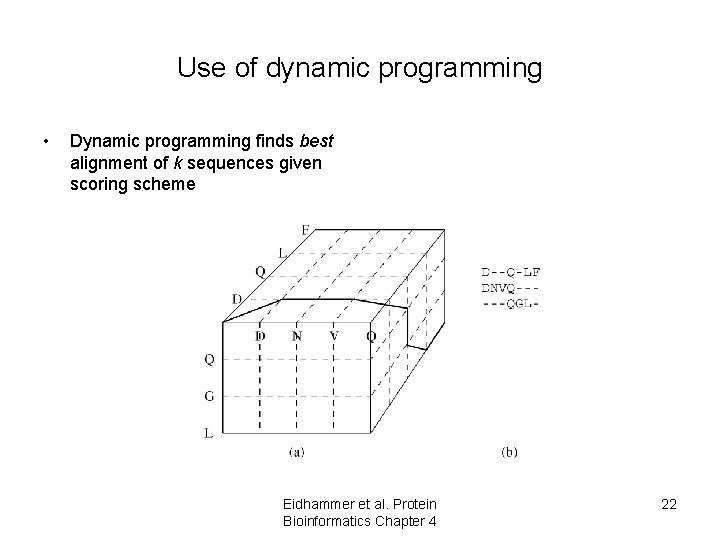
Use of dynamic programming • Dynamic programming finds best alignment of k sequences given scoring scheme Eidhammer et al. Protein Bioinformatics Chapter 4 22

Eidhammer et al. Protein Bioinformatics Chapter 4 23
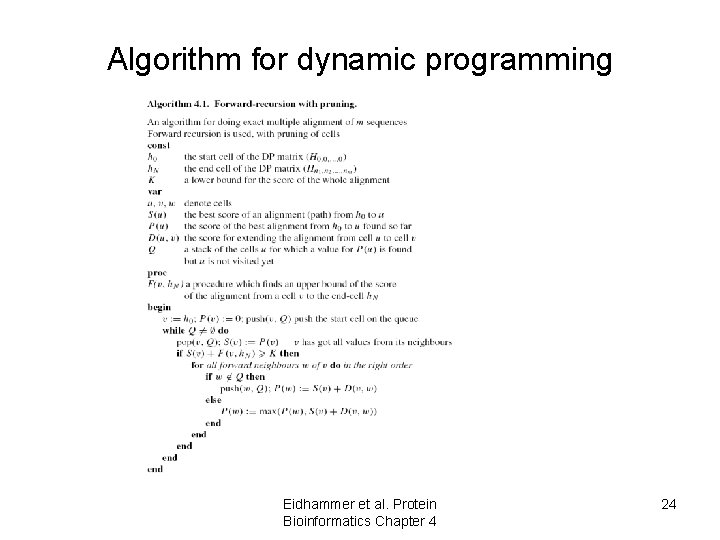
Algorithm for dynamic programming Eidhammer et al. Protein Bioinformatics Chapter 4 24
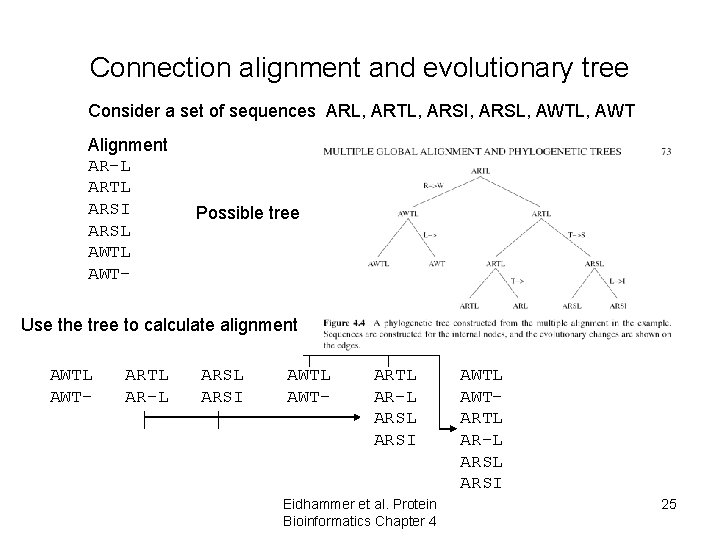
Connection alignment and evolutionary tree Consider a set of sequences ARL, ARTL, ARSI, ARSL, AWT Alignment AR-L ARTL ARSI ARSL AWT- Possible tree Use the tree to calculate alignment AWTL AWT- ARTL AR-L ARSL ARSI Eidhammer et al. Protein Bioinformatics Chapter 4 AWTL AWTARTL AR-L ARSI 25
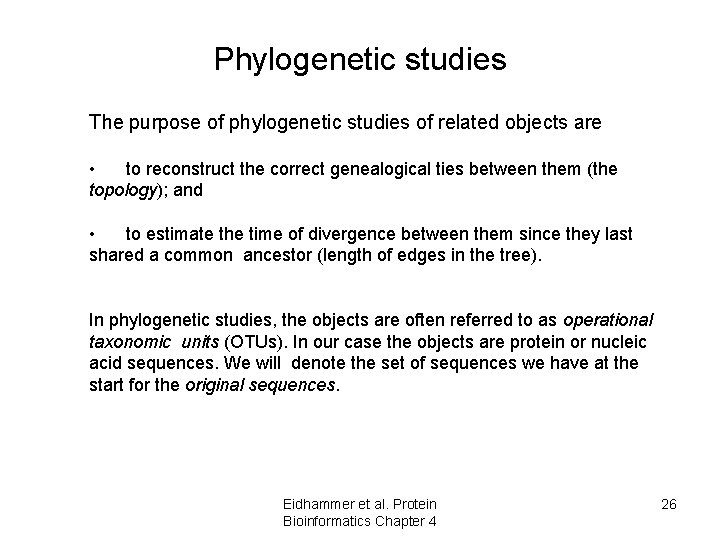
Phylogenetic studies The purpose of phylogenetic studies of related objects are • to reconstruct the correct genealogical ties between them (the topology); and • to estimate the time of divergence between them since they last shared a common ancestor (length of edges in the tree). In phylogenetic studies, the objects are often referred to as operational taxonomic units (OTUs). In our case the objects are protein or nucleic acid sequences. We will denote the set of sequences we have at the start for the original sequences. Eidhammer et al. Protein Bioinformatics Chapter 4 26

Phylogenetic studies Eidhammer et al. Protein Bioinformatics Chapter 4 27
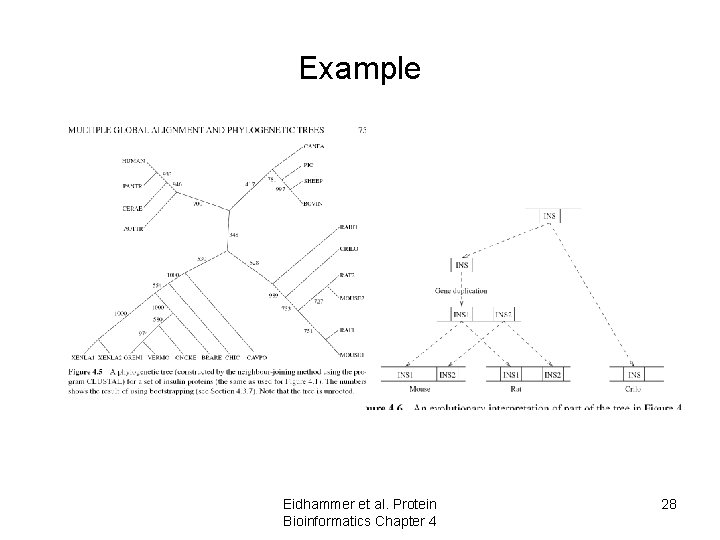
Example Eidhammer et al. Protein Bioinformatics Chapter 4 28
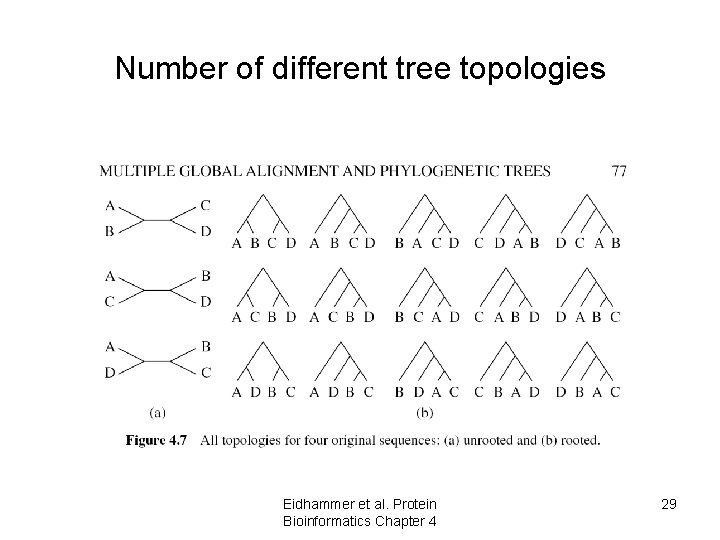
Number of different tree topologies Eidhammer et al. Protein Bioinformatics Chapter 4 29
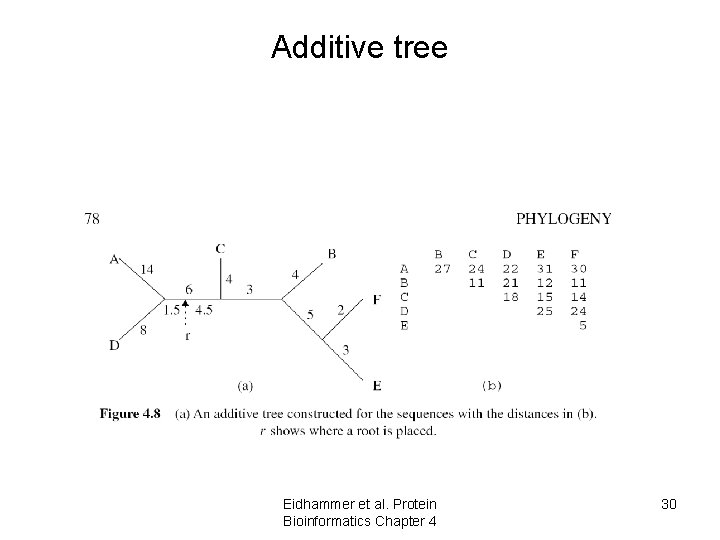
Additive tree Eidhammer et al. Protein Bioinformatics Chapter 4 30
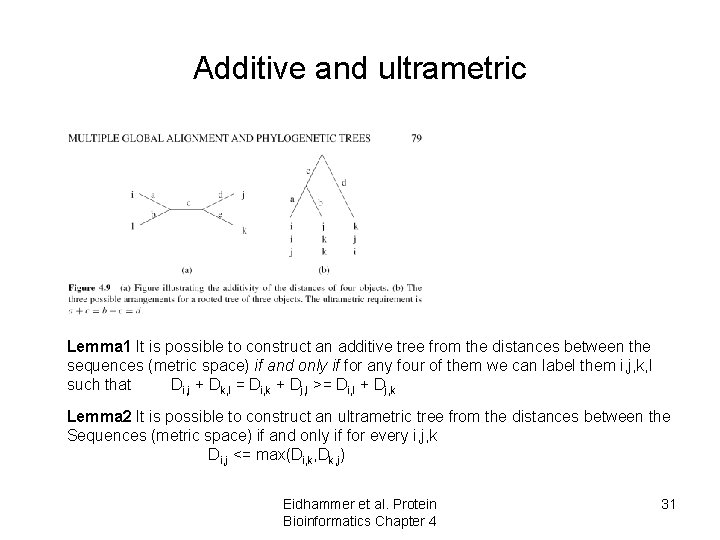
Additive and ultrametric Lemma 1 It is possible to construct an additive tree from the distances between the sequences (metric space) if and only if for any four of them we can label them i, j, k, l such that Di, j + Dk, l = Di, k + Dj, l >= Di, l + Dj, k Lemma 2 It is possible to construct an ultrametric tree from the distances between the Sequences (metric space) if and only if for every i, j, k Di, j <= max(Di, k, Dk, j) Eidhammer et al. Protein Bioinformatics Chapter 4 31
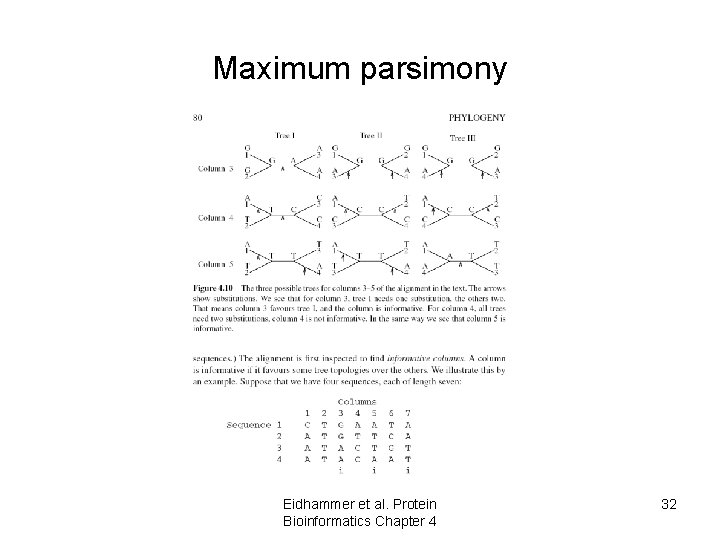
Maximum parsimony Eidhammer et al. Protein Bioinformatics Chapter 4 32

Parvis gruppering Eidhammer et al. Protein Bioinformatics Chapter 4 33
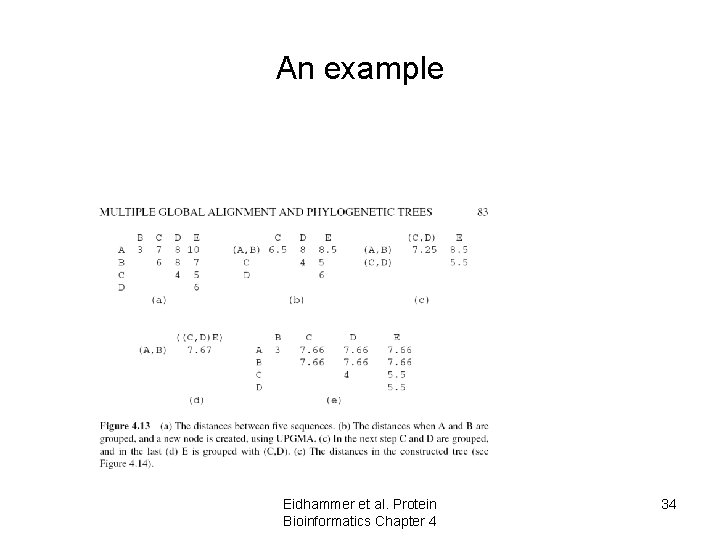
An example Eidhammer et al. Protein Bioinformatics Chapter 4 34
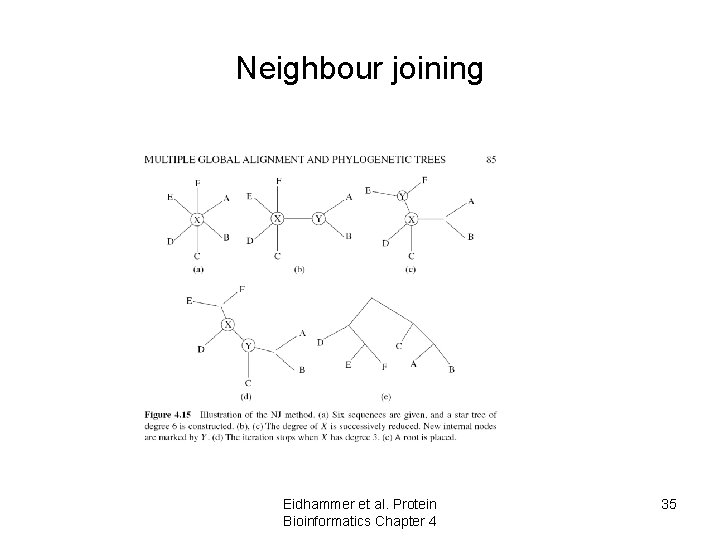
Neighbour joining Eidhammer et al. Protein Bioinformatics Chapter 4 35
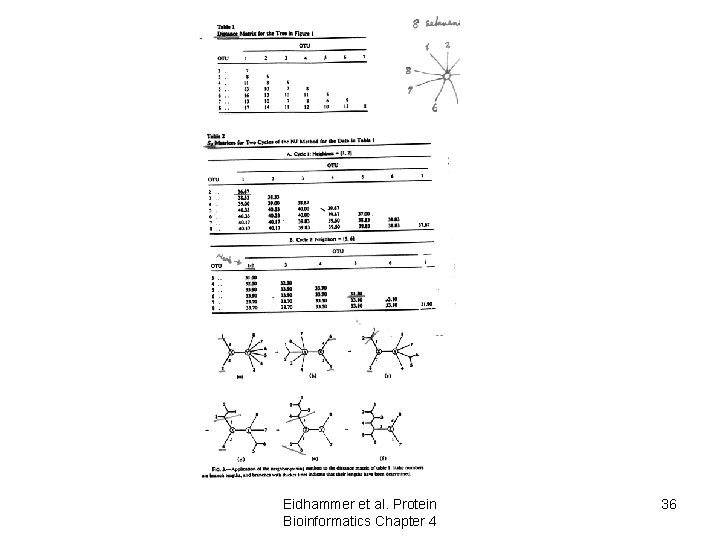
Eidhammer et al. Protein Bioinformatics Chapter 4 36

Bootstrapping Eidhammer et al. Protein Bioinformatics Chapter 4 37

Eidhammer et al. Protein Bioinformatics Chapter 4 38
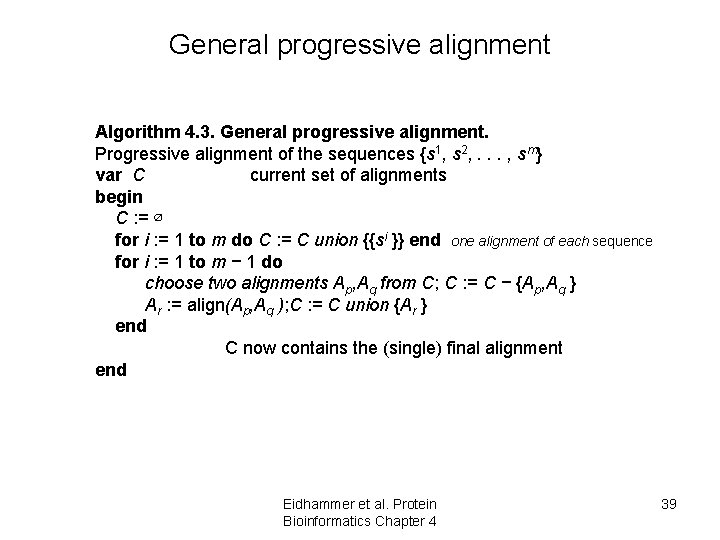
General progressive alignment Algorithm 4. 3. General progressive alignment. Progressive alignment of the sequences {s 1, s 2, . . . , sm} var C current set of alignments begin C : = ∅ for i : = 1 to m do C : = C union {{si }} end one alignment of each sequence for i : = 1 to m − 1 do choose two alignments Ap, Aq from C; C : = C − {Ap, Aq } Ar : = align(Ap, Aq ); C : = C union {Ar } end C now contains the (single) final alignment end Eidhammer et al. Protein Bioinformatics Chapter 4 39

Eidhammer et al. Protein Bioinformatics Chapter 4 40

Eidhammer et al. Protein Bioinformatics Chapter 4 41

Eidhammer et al. Protein Bioinformatics Chapter 4 42

Clustering philosophy Join the two groups with highest pairwise score. 1. 2. 3. 4. Average scoring method: find average score over all pasirs in the two groups Maximum scoring method: find maximum score over all pairs in the two groups (needs only one high-scoring pair) Minimum (complete) scoring method: find minimum scoring over all pairs (all pairs are taken into account) Special scoring method Eidhammer et al. Protein Bioinformatics Chapter 4 43

Eidhammer et al. Protein Bioinformatics Chapter 4 44
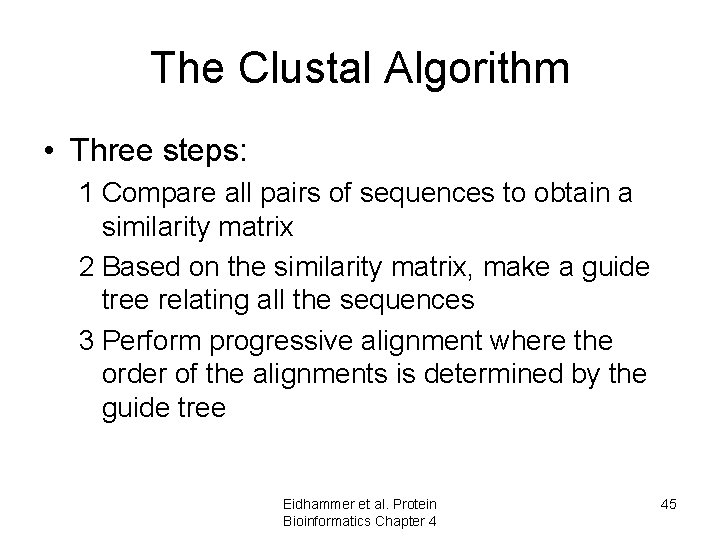
The Clustal Algorithm • Three steps: 1 Compare all pairs of sequences to obtain a similarity matrix 2 Based on the similarity matrix, make a guide tree relating all the sequences 3 Perform progressive alignment where the order of the alignments is determined by the guide tree Eidhammer et al. Protein Bioinformatics Chapter 4 45
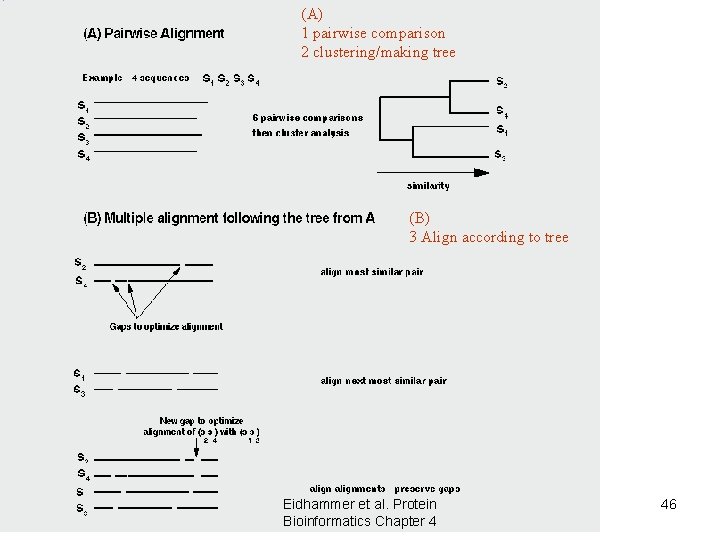
(A) 1 pairwise comparison 2 clustering/making tree (B) 3 Align according to tree Eidhammer et al. Protein Bioinformatics Chapter 4 46
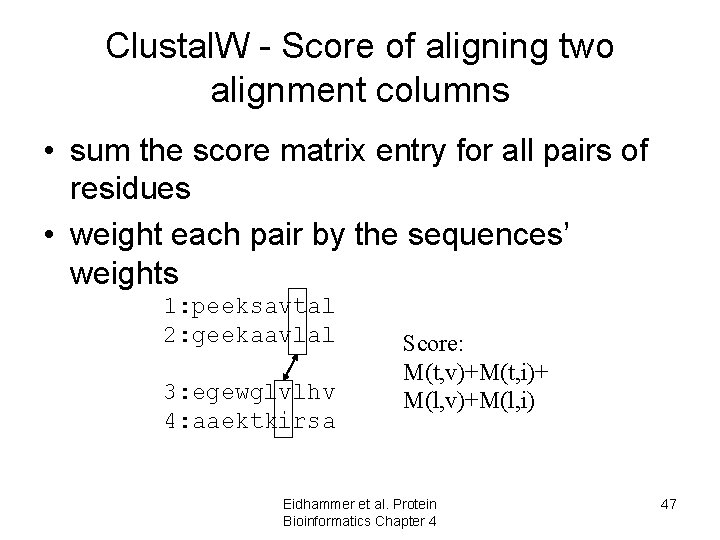
Clustal. W - Score of aligning two alignment columns • sum the score matrix entry for all pairs of residues • weight each pair by the sequences’ weights 1: peeksavtal 2: geekaavlal 3: egewglvlhv 4: aaektkirsa Score: M(t, v)+M(t, i)+ M(l, v)+M(l, i) Eidhammer et al. Protein Bioinformatics Chapter 4 47
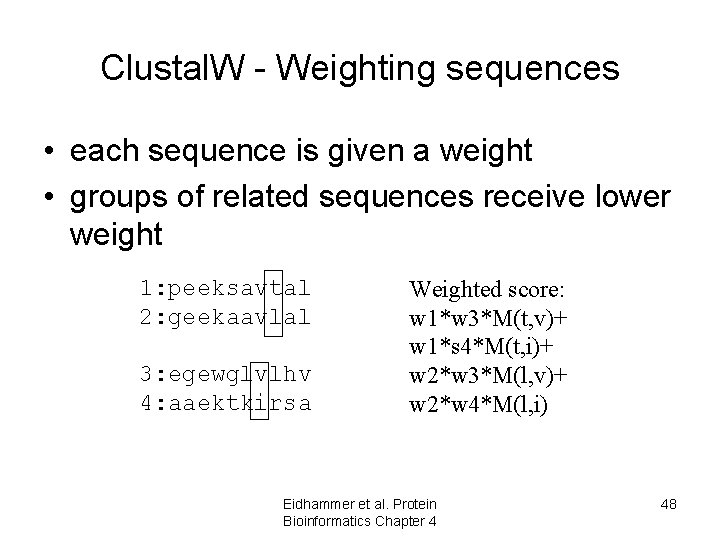
Clustal. W - Weighting sequences • each sequence is given a weight • groups of related sequences receive lower weight 1: peeksavtal 2: geekaavlal 3: egewglvlhv 4: aaektkirsa Weighted score: w 1*w 3*M(t, v)+ w 1*s 4*M(t, i)+ w 2*w 3*M(l, v)+ w 2*w 4*M(l, i) Eidhammer et al. Protein Bioinformatics Chapter 4 48
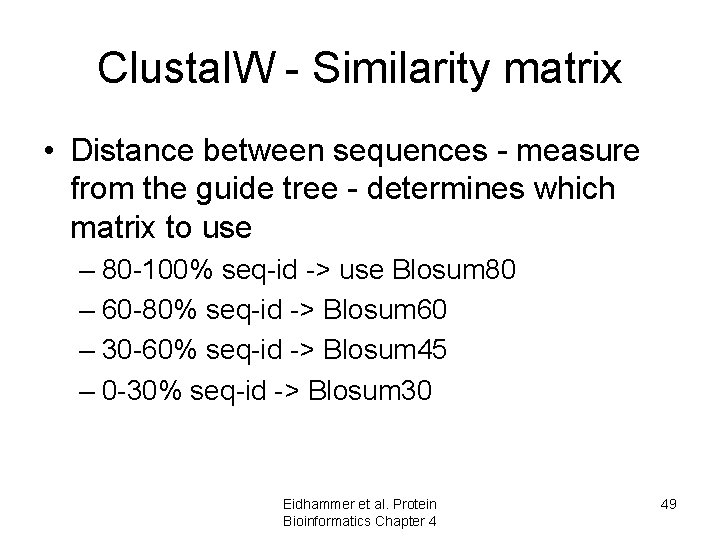
Clustal. W - Similarity matrix • Distance between sequences - measure from the guide tree - determines which matrix to use – 80 -100% seq-id -> use Blosum 80 – 60 -80% seq-id -> Blosum 60 – 30 -60% seq-id -> Blosum 45 – 0 -30% seq-id -> Blosum 30 Eidhammer et al. Protein Bioinformatics Chapter 4 49

Clustal. W - Gap penalties • Initial gap penalty – GOP GTEAKLIVLMANE GA-----KL • Gap extension penalty – GEP Penalty: GOP+8*GEP Eidhammer et al. Protein Bioinformatics Chapter 4 50

Clustal. W Modifications of gap penalty • Position specific penalty – gap at position • yes -> lower GOP • no, but gap within 8 residues -> increase GOP – hydrophilic residues • lower GOP Eidhammer et al. Protein Bioinformatics Chapter 4 51
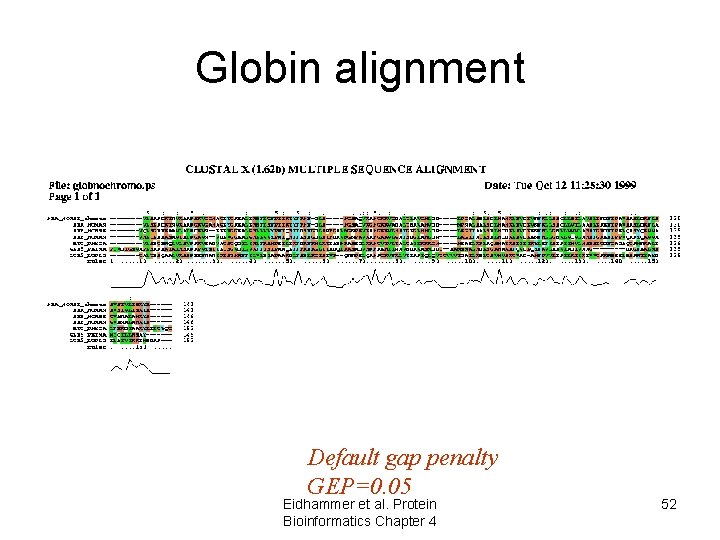
Globin alignment Default gap penalty GEP=0. 05 Eidhammer et al. Protein Bioinformatics Chapter 4 52
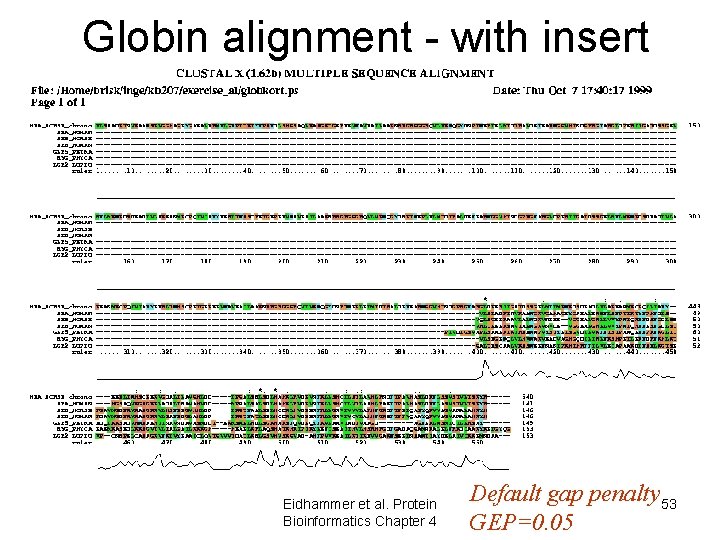
Globin alignment - with insert Eidhammer et al. Protein Bioinformatics Chapter 4 Default gap penalty 53 GEP=0. 05
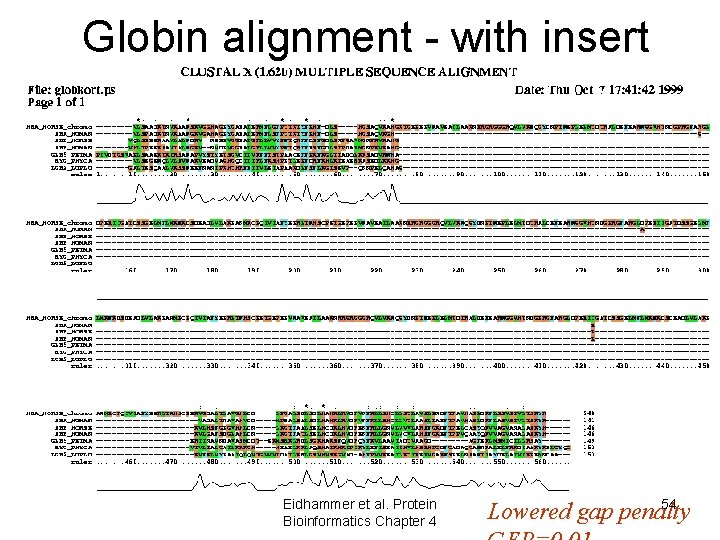
Globin alignment - with insert Eidhammer et al. Protein Bioinformatics Chapter 4 54 Lowered gap penalty
Clustal. W - summary • Does not use a score for the final alignment • Each pairwise alignment is done using dynamic programming • Heuristics (e. g. , gap-penalty modifications) are used - tailored to globular proteins • Graphical version: Clustal. X Eidhammer et al. Protein Bioinformatics Chapter 4 55
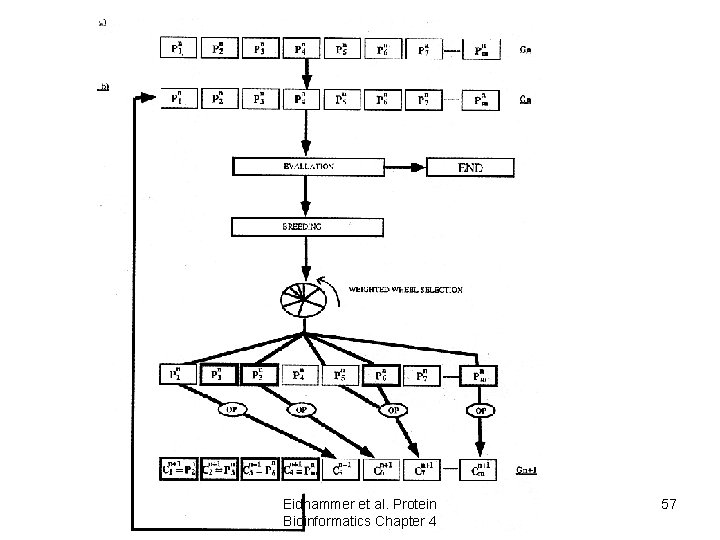
SAGA: Sequence Alignment by Genetic Algorithm • An “objective function” is used to score the alignments • An alignment is represented as a bit string • A population of alignment is “evolved” • Alignments can be combined (crossover) • Alignments can be etmutated Eidhammer al. Protein Bioinformatics Chapter 4 56
Eidhammer et al. Protein Bioinformatics Chapter 4 57
Evaluation of Alignment Methods • Align set of protein sequences where the structures are known (at least for some proteins) • Align the protein structures • Identify “motifs” from the structure alignment • Check if sequence alignment has correctly aligned motifs Eidhammer et al. Protein Bioinformatics Chapter 4 58
Alignments are important • Basis for other analyses – structure prediction – phylogeny – experiments • PCR primer identification • site directed mutagenesis • . . . – identification of motifs Eidhammer et al. Protein Bioinformatics Chapter 4 59
Open Problems space for improvements! • Good scoring function for alignments – identify well aligned regions • Efficient algorithms • Resolving repeat structure, domain movements etc. • Incorporating external information Eidhammer et al. Protein Bioinformatics Chapter 4 60
Future development • More sequences – More families, but not so many – More densely populated families – “Easier” alignment problem – Identify more ancient relationships (superfamilies) • More structures – more sequences can be “threaded” – alignments help Eidhammer et al. Protein Bioinformatics Chapter 4 61
- Slides: 61