Molecular Similarity Molecular Descriptors for Drug Design N









![Protein fingerprint — Mihaly Mezei FP 0 ij= sign {[r(Oi)-r(Ci)]. [r(Cj)-r(Ci)]} FP 1 ij= Protein fingerprint — Mihaly Mezei FP 0 ij= sign {[r(Oi)-r(Ci)]. [r(Cj)-r(Ci)]} FP 1 ij=](https://slidetodoc.com/presentation_image_h/e99d84f7752532567d087e571c37c3ef/image-48.jpg)

- Slides: 50

Molecular Similarity & Molecular Descriptors for Drug Design N. Sukumar Center for Biotechnology & Interdisciplinary Studies Rensselaer Polytechnic Institute

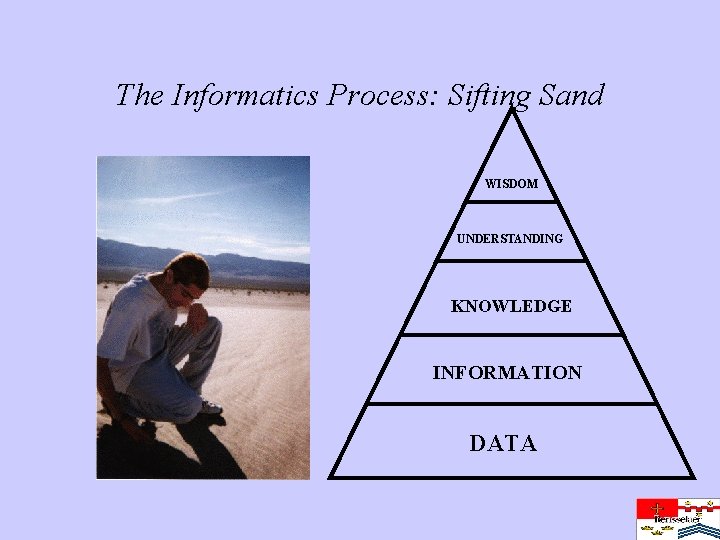
The Informatics Process: Sifting Sand WISDOM UNDERSTANDING KNOWLEDGE INFORMATION DATA

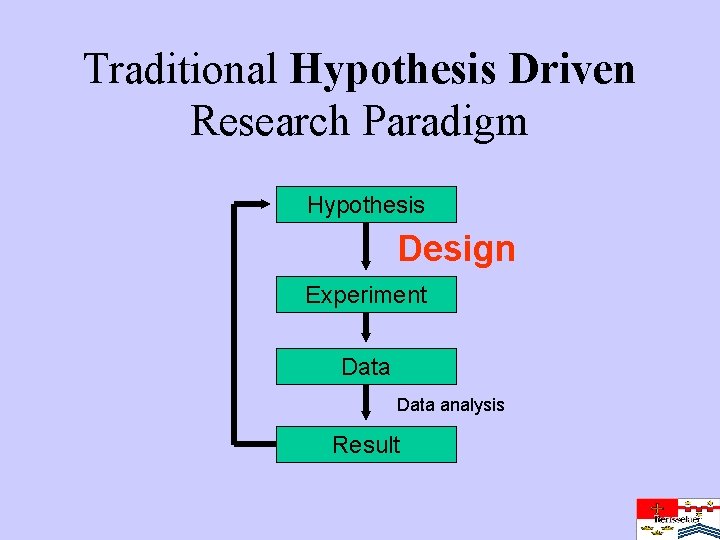
Traditional Hypothesis Driven Research Paradigm Hypothesis Design Experiment Data analysis Result

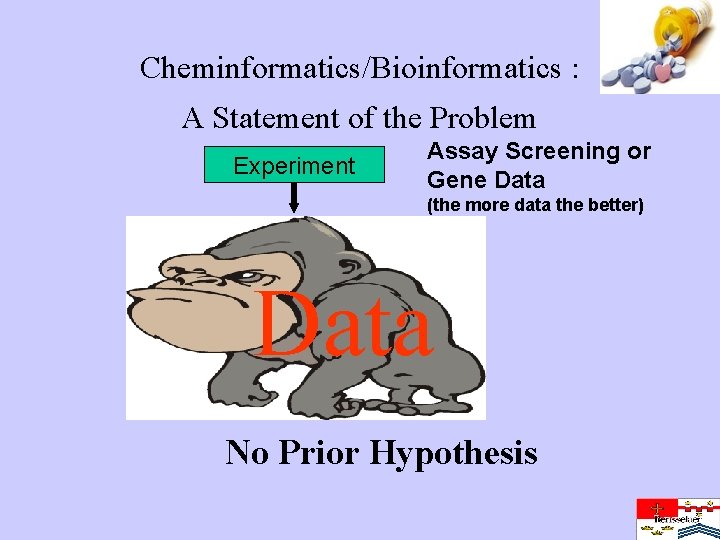
Cheminformatics/Bioinformatics : A Statement of the Problem Experiment Assay Screening or Gene Data (the more data the better) Data No Prior Hypothesis

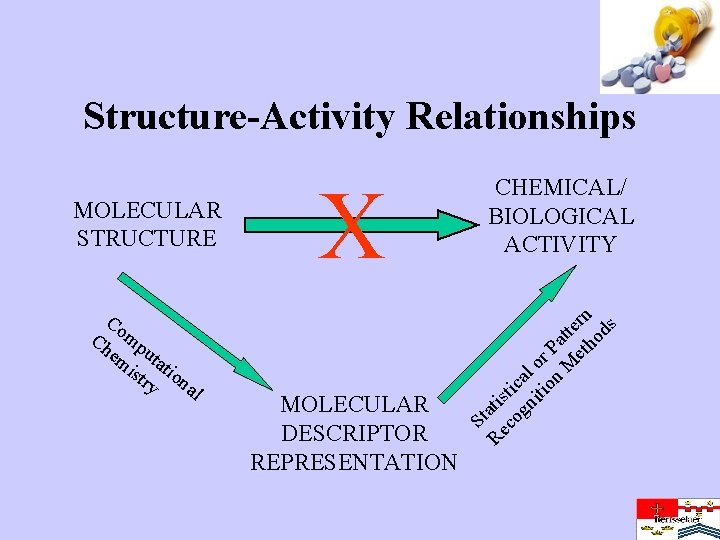
Structure-Activity Relationships Co Ch mp em uta ist tio ry na l X MOLECULAR DESCRIPTOR REPRESENTATION St Re atis co tic gn al iti or on P M att et ern ho ds MOLECULAR STRUCTURE CHEMICAL/ BIOLOGICAL ACTIVITY

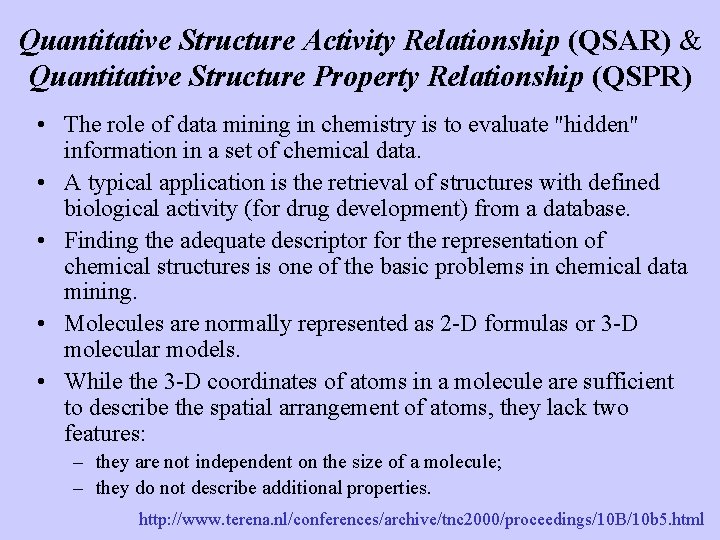
Quantitative Structure Activity Relationship (QSAR) & Quantitative Structure Property Relationship (QSPR) • The role of data mining in chemistry is to evaluate "hidden" information in a set of chemical data. • A typical application is the retrieval of structures with defined biological activity (for drug development) from a database. • Finding the adequate descriptor for the representation of chemical structures is one of the basic problems in chemical data mining. • Molecules are normally represented as 2 -D formulas or 3 -D molecular models. • While the 3 -D coordinates of atoms in a molecule are sufficient to describe the spatial arrangement of atoms, they lack two features: – they are not independent on the size of a molecule; – they do not describe additional properties. http: //www. terena. nl/conferences/archive/tnc 2000/proceedings/10 B/10 b 5. html

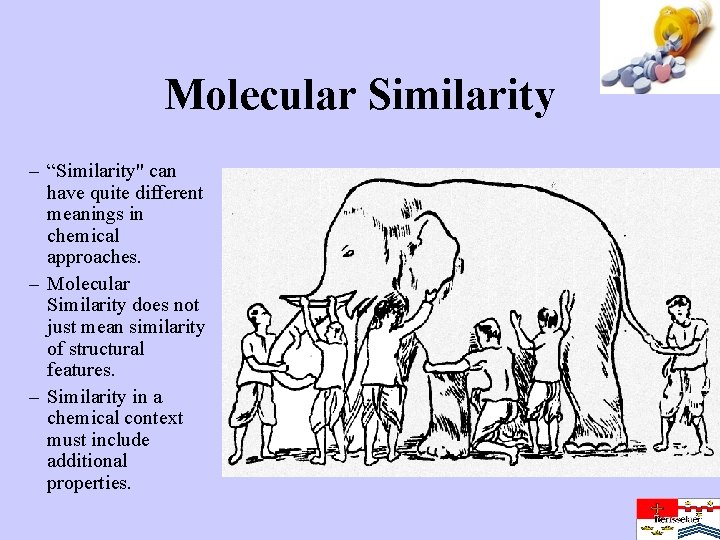
Molecular Similarity – “Similarity" can have quite different meanings in chemical approaches. – Molecular Similarity does not just mean similarity of structural features. – Similarity in a chemical context must include additional properties.

It was six men of Indostan To learning much inclined, Who went to see the Elephant (Though all of them were blind), That each by observation Might satisfy his mind The First approached the Elephant, And happening to fall Against his broad and sturdy side, At once began to bawl: “God bless me! but the Elephant Is very like a wall!” The Second, feeling of the tusk, Cried, “Ho! what have we here So very round and smooth and sharp? To me ’tis mighty clear This wonder of an Elephant Is very like a spear!” The Third approached the animal, And happening to take The squirming trunk within his hands, Thus boldly up and spake: “I see, ” quoth he, “the Elephant Is very like a snake!” The Fourth reached out an eager hand, And felt about the knee. “What most this wondrous beast is like Is mighty plain, ” quoth he; “ ‘Tis clear enough the Elephant Is very like a tree!” The Fifth, who chanced to touch the ear, Said: “E’en the blindest man Can tell what this resembles most; Deny the fact who can This marvel of an Elephant Is very like a fan!” The Sixth no sooner had begun About the beast to grope, Than, seizing on the swinging tail That fell within his scope, “I see, ” quoth he, “the Elephant Is very like a rope!” And so these men of Indostan Disputed loud and long, Each in his own opinion Exceeding stiff and strong, Though each was partly in the right, And all were in the wrong! - John Godfrey Saxe (1816 -1887)

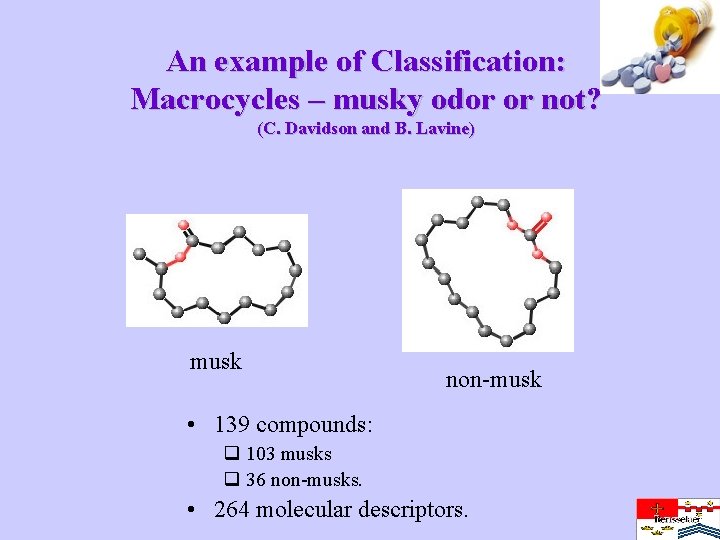
An example of Classification: Macrocycles – musky odor or not? (C. Davidson and B. Lavine) musk non-musk • 139 compounds: q 103 musks q 36 non-musks. • 264 molecular descriptors.

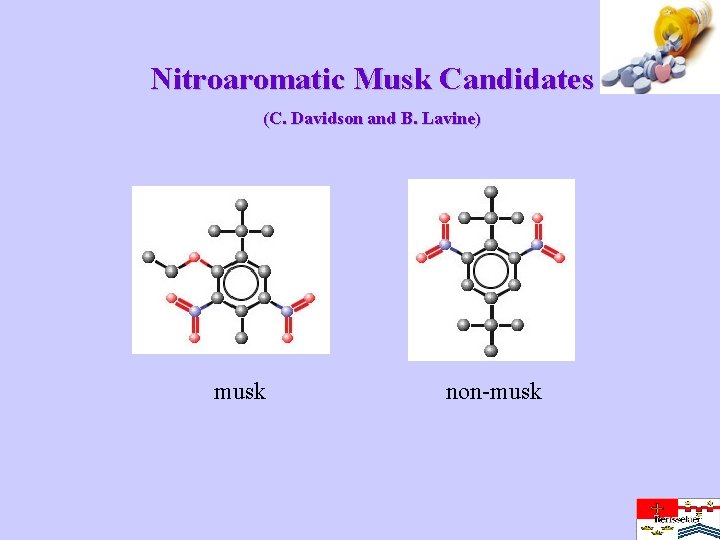
Nitroaromatic Musk Candidates (C. Davidson and B. Lavine) musk non-musk

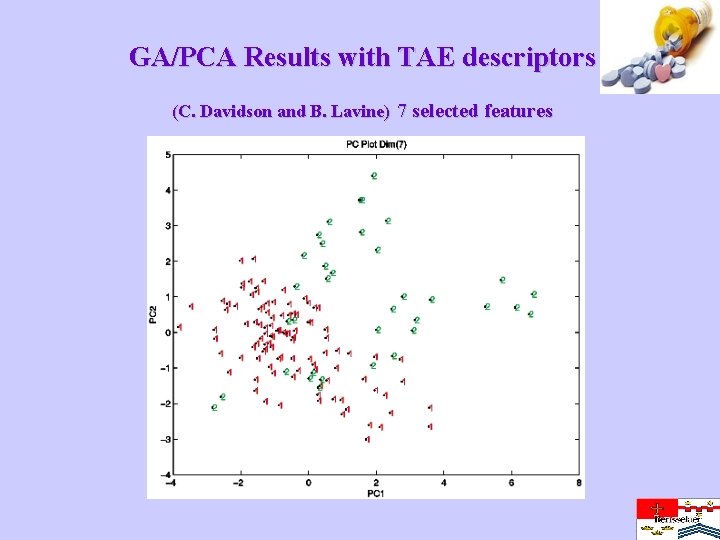
GA/PCA Results with TAE descriptors (C. Davidson and B. Lavine) 7 selected features

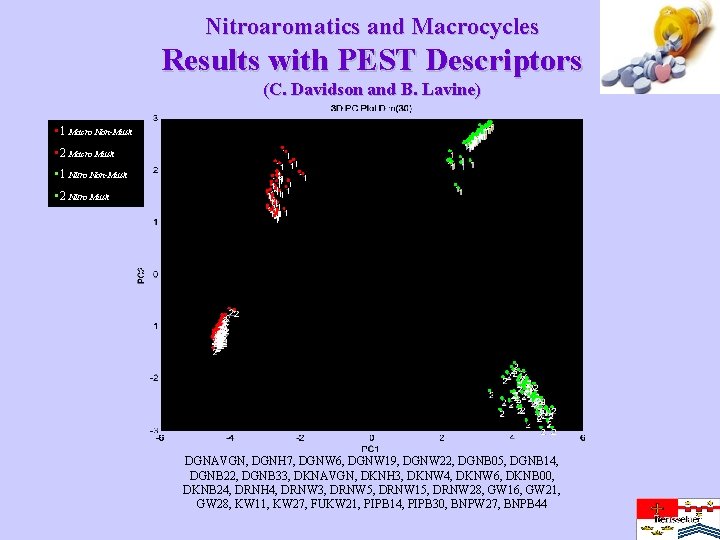
Nitroaromatics and Macrocycles Results with PEST Descriptors (C. Davidson and B. Lavine) • 1 Macro Non-Musk • 2 Macro Musk • 1 Nitro Non-Musk • 2 Nitro Musk DGNAVGN, DGNH 7, DGNW 6, DGNW 19, DGNW 22, DGNB 05, DGNB 14, DGNB 22, DGNB 33, DKNAVGN, DKNH 3, DKNW 4, DKNW 6, DKNB 00, DKNB 24, DRNH 4, DRNW 3, DRNW 5, DRNW 15, DRNW 28, GW 16, GW 21, GW 28, KW 11, KW 27, FUKW 21, PIPB 14, PIPB 30, BNPW 27, BNPB 44

ADMET Property Prediction: Challenges in Medicinal Chemistry Multipleparameter optimization of lead structures • Other parameters: patent position, chemical synthesis • The greatest hurdle : ADMET properties.

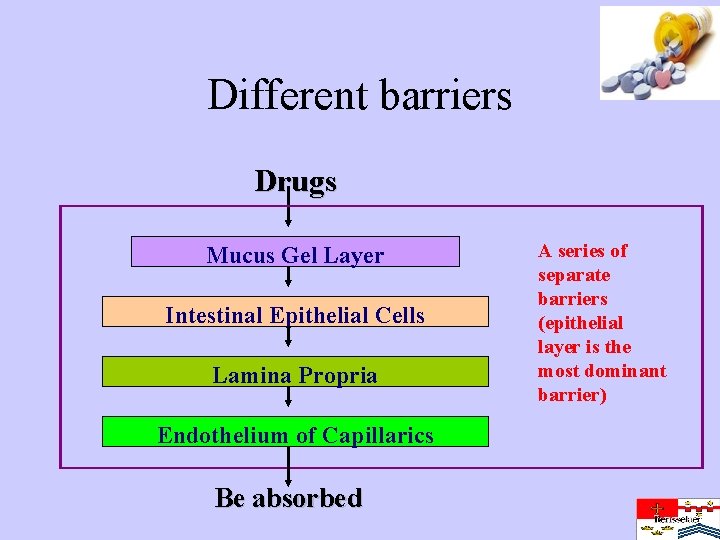
Different barriers Drugs Mucus Gel Layer Intestinal Epithelial Cells Lamina Propria Endothelium of Capillarics Be absorbed A series of separate barriers (epithelial layer is the most dominant barrier)

Motivation • Introduction of a new drug into the market is often the culmination of a long and arduous process of laboratory experimentation, lead compound discovery, animal testing and pre-clinical and clinical trials. • This process, from hit to lead to marketable drug, is typically as long as 10 -15 years • In silico drug discovery: – find a correlation between molecular structure and biological activity – now any number of compounds, including those not yet synthesized, can be virtually screened on the computer to select structures with the desired properties. • Virtual ADME/Toxicological screening can weed out compounds with adverse side effects, identifying the “losers” early on in the game. • The most promising compounds can then be chosen for laboratory synthesis and pre-clinical testing – conserving resources cheaper medicines – accelerating the process of drug discovery.

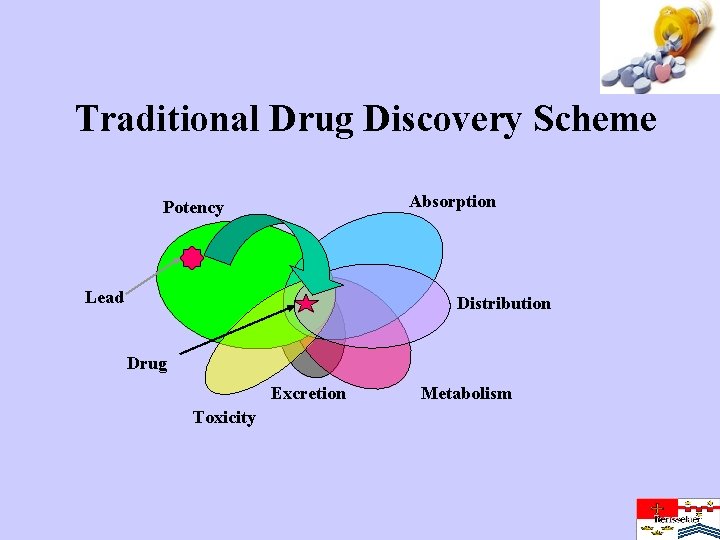
Traditional Drug Discovery Scheme Absorption Potency Lead Distribution Drug Excretion Toxicity Metabolism

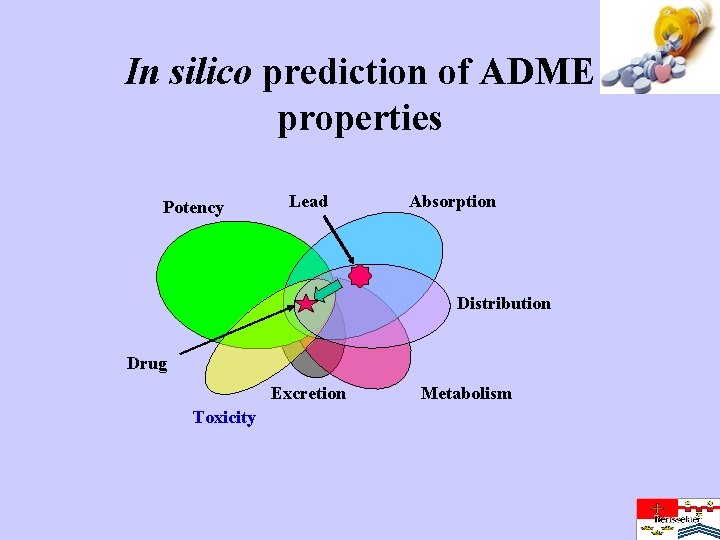
In silico prediction of ADME properties Potency Lead Absorption Distribution Drug Excretion Toxicity Metabolism

Computational ADME-Tox models for drug discovery • • Solubility Absorption Mutagenicity Bioavailability Metabolic stability Blood-brain barrier permeability Cardiac toxicity (h. ERG) Plasma protein binding

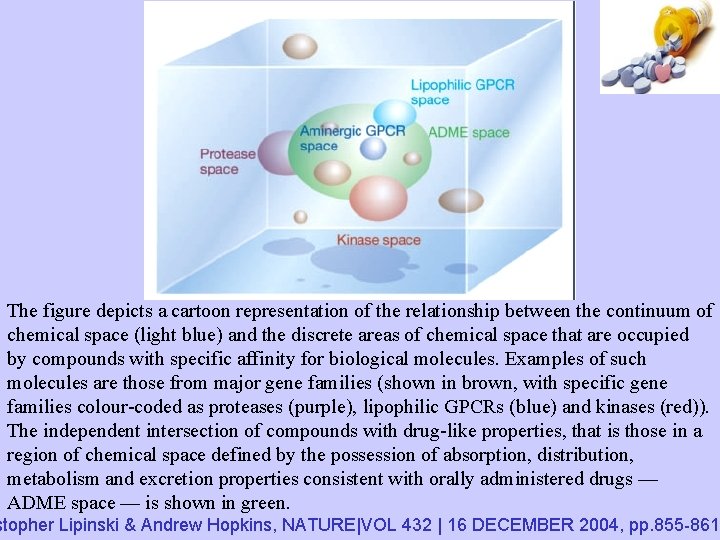
The figure depicts a cartoon representation of the relationship between the continuum of chemical space (light blue) and the discrete areas of chemical space that are occupied by compounds with specific affinity for biological molecules. Examples of such molecules are those from major gene families (shown in brown, with specific gene families colour-coded as proteases (purple), lipophilic GPCRs (blue) and kinases (red)). The independent intersection of compounds with drug-like properties, that is those in a region of chemical space defined by the possession of absorption, distribution, metabolism and excretion properties consistent with orally administered drugs — ADME space — is shown in green. stopher Lipinski & Andrew Hopkins, NATURE|VOL 432 | 16 DECEMBER 2004, pp. 855 -861

Descriptors from Molecular Electronic Properties

Molecular Representations

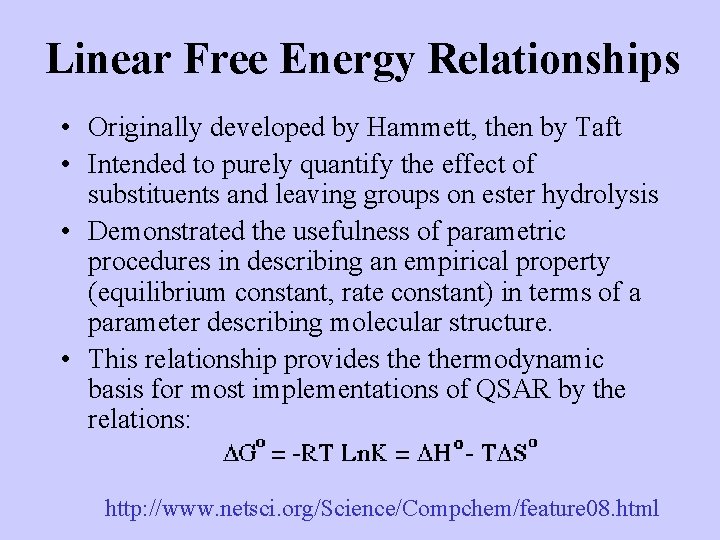
Linear Free Energy Relationships • Originally developed by Hammett, then by Taft • Intended to purely quantify the effect of substituents and leaving groups on ester hydrolysis • Demonstrated the usefulness of parametric procedures in describing an empirical property (equilibrium constant, rate constant) in terms of a parameter describing molecular structure. • This relationship provides thermodynamic basis for most implementations of QSAR by the relations: http: //www. netsci. org/Science/Compchem/feature 08. html

Quantitative Structure-Activity Relationships (QSAR) • QSAR was a natural extension of the LFER approach, with a biological activity correlated against a series of parameters that described the structure of a molecule. • The most well known and most used descriptor in QSAR has been the LOG (Octanol/Water) partition coefficient (usually referred to as LOG P or LOG P[o/w]). LOG P has been very useful in correlating a wide range of activities due to its excellent modeling of the transport across the blood/brain barrier. • Unfortunately, many regressions do not work well for LOG P, usually because other effects are important, such as steric and electronic effects. • Therefore, many other descriptors have been used in QSAR in addition to LOG P to incorporate these additional effects.

• Number of aromatic atoms • Number of heavy atoms ® Chemical Computing Group Inc. • Number of hydrogen atoms • Number of boron atoms “ 2 -D” Molecular Descriptors can be calculated from the • Number of carbon atoms • Number of nitrogen atoms connection table (with no dependence on conformation): • Number of oxygen atoms • Sum of the atomic polarizabilities • Number of fluorine atoms – Physical Properties • Molecular mass density • Number of phosphorus atoms • Total charge of the molecule – Subdivided Surface Area Descriptors • Number of sulfur atoms • Molecular refractivity – Atom Counts and Bond Counts • Molecular weight. • Number of chlorine atoms • Water accessible surface area of all atoms with positive partial charge • Number of bromine atoms • Log of the octanol/water partition • Water accessible surface area of all atoms with negative partial charge – Connectivity and Shape Indices • Number of iodine atoms coefficient • Water accessible surface area of all hydrophobic atoms • Number of rotatable single bonds – Adjacency and Distance Matrix Descriptors • Water accessible surface area of all polar atoms • Number of aromatic bonds • Positive charge weighted surface area • Number of hydrogen bond acceptor atoms – Pharmacophore Feature Descriptors • Number of bonds • Negative charge weighted surface area • Number of acidic atoms • Number of double bonds – Partial Charge Descriptors • Total positive partial charge • Number of basic atoms • Total negative partial charge • Number of rotatable bonds • Number of hydrogen bond donor atoms • Total positive van der Waals surface area “ 3 -D” Descriptors depend on molecular coordinates: • Fraction of rotatable bonds • Angle bend potential energy • Number of hydrophobic atoms • Total negative van der Waals surface area • Number of single bonds • Electrostatic component of the potential energy – Potential Energy Descriptors • Fractional positive polar van der Waals surface area • Number of triple bonds • Out-of-plane potential energy • Water accessible surface area • Fractional negative polar van der Waals surface area – Surface Area, Volume and Shape Descriptors • Number of chiral centers • Solvation energy • Globularity • Number of O and N atoms • Bond stretch potential energy – Conformation Dependent Charge Descriptors • Principal moment of inertia • Number of OH and NH groups • Local strain energy • Radius of gyration • Number of rings • Torsion potential energy • van der Waals surface area MOE Descriptors • •

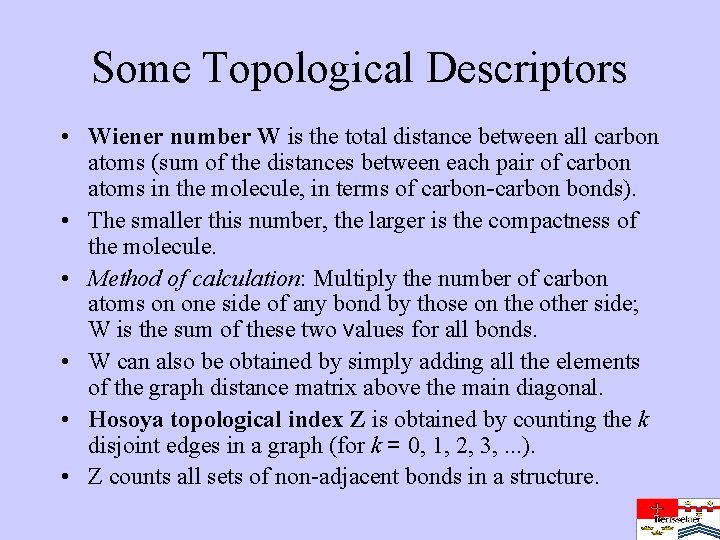
Some Topological Descriptors • Wiener number W is the total distance between all carbon atoms (sum of the distances between each pair of carbon atoms in the molecule, in terms of carbon-carbon bonds). • The smaller this number, the larger is the compactness of the molecule. • Method of calculation: Multiply the number of carbon atoms on one side of any bond by those on the other side; W is the sum of these two values for all bonds. • W can also be obtained by simply adding all the elements of the graph distance matrix above the main diagonal. • Hosoya topological index Z is obtained by counting the k disjoint edges in a graph (for k = 0, 1, 2, 3, . . . ). • Z counts all sets of non-adjacent bonds in a structure.

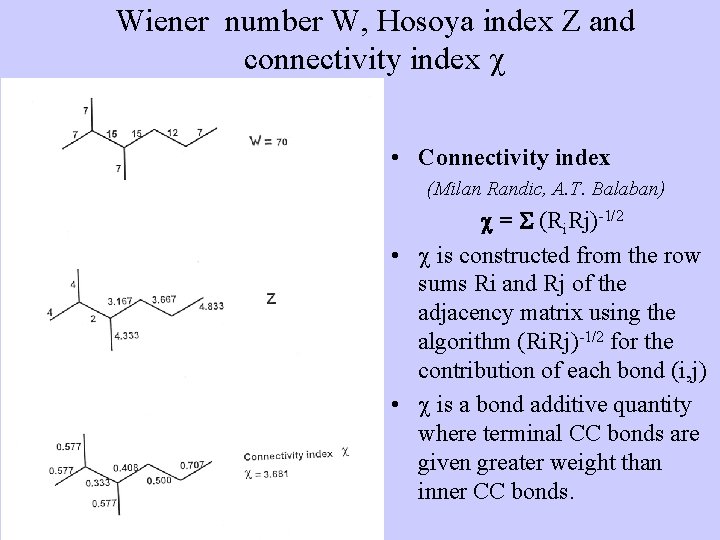
Wiener number W, Hosoya index Z and connectivity index • Connectivity index (Milan Randic, A. T. Balaban) = (Ri. Rj)-1/2 • is constructed from the row sums Ri and Rj of the adjacency matrix using the algorithm (Ri. Rj)-1/2 for the contribution of each bond (i, j) • is a bond additive quantity where terminal CC bonds are given greater weight than inner CC bonds.

Quantum chemical Electron Density Derived descriptors – The wave function given by solution of the Schrödinger equation H = E contains all information about the molecule. – “All science is either physics or stamp collecting” — Ernest Rutherford (Nobel Prize in Chemistry, 1908) – BUT: (r 1, r 2, r 3, …) is a function of the coordinates of all the electrons (and nuclei) in the molecule! – “The fundamental laws necessary for the mathematical treatment of a large part of physics and the whole of chemistry are thus completely known, and the difficulty lies only in the fact that application of these laws leads to equations that are too complex to be solved. ” — Paul Dirac (1902 - 1984)

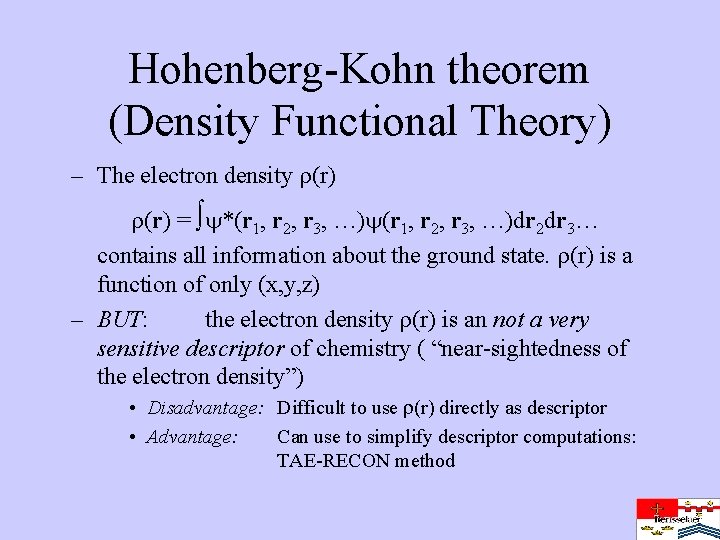
Hohenberg-Kohn theorem (Density Functional Theory) – The electron density (r) = *(r 1, r 2, r 3, …)dr 2 dr 3… contains all information about the ground state. (r) is a function of only (x, y, z) – BUT: the electron density (r) is an not a very sensitive descriptor of chemistry ( “near-sightedness of the electron density”) • Disadvantage: Difficult to use (r) directly as descriptor • Advantage: Can use to simplify descriptor computations: TAE-RECON method

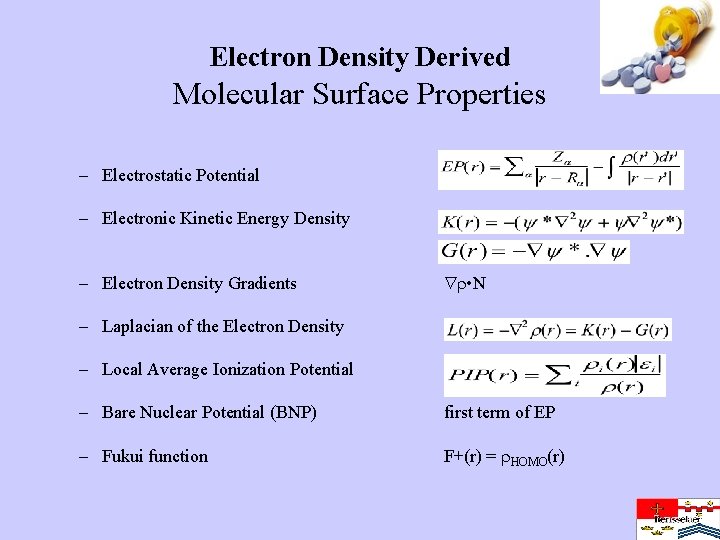
Electron Density Derived Molecular Surface Properties – Electrostatic Potential – Electronic Kinetic Energy Density – Electron Density Gradients • N – Laplacian of the Electron Density – Local Average Ionization Potential – Bare Nuclear Potential (BNP) first term of EP – Fukui function F+(r) = HOMO(r)

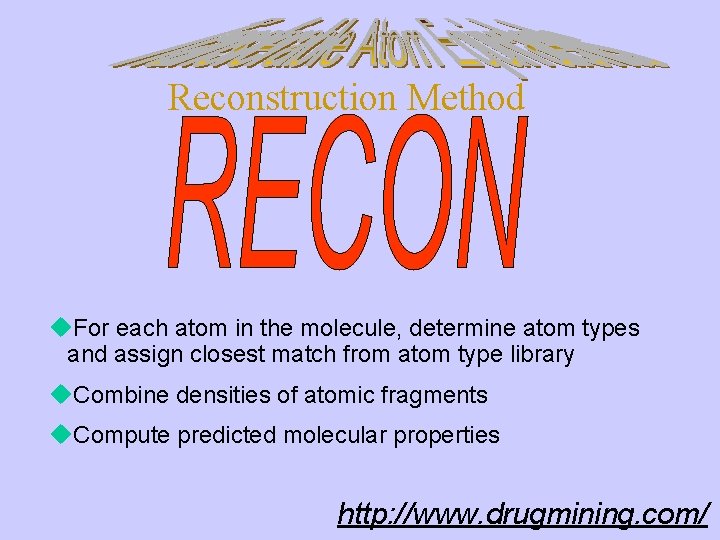
Reconstruction Method u Algorithm for rapid reconstruction of molecular charge densities and molecular electronic properties u Based on topological quantum theory of Atoms In Molecules u Employs a library of atomic charge density fragments corresponding to structurally distinct atom types u Associated with each atomic charge density fragment in the library is a data file which contains atomic charge density-based descriptors encoding electronic and structural information relevant to the chemistry of intermolecular interactions. http: //www. drugmining. com/

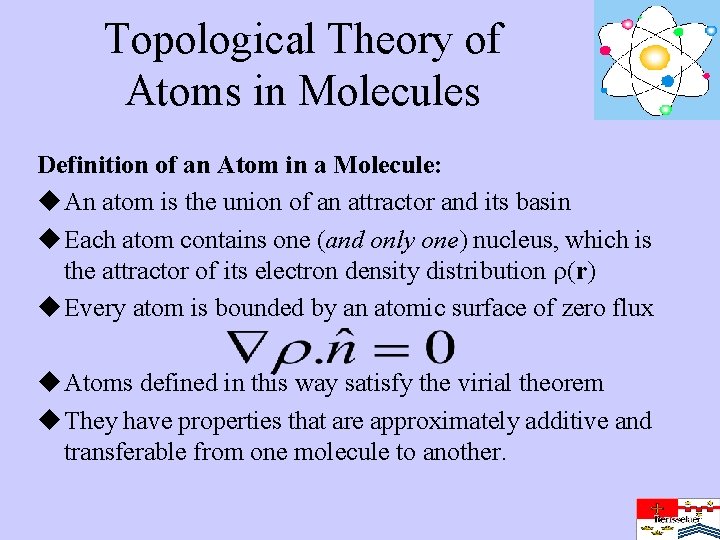
Topological Theory of Atoms in Molecules Definition of an Atom in a Molecule: u An atom is the union of an attractor and its basin u Each atom contains one (and only one) nucleus, which is the attractor of its electron density distribution (r) u Every atom is bounded by an atomic surface of zero flux u Atoms defined in this way satisfy the virial theorem u They have properties that are approximately additive and transferable from one molecule to another.

Reconstruction Method u. For each atom in the molecule, determine atom types and assign closest match from atom type library u. Combine densities of atomic fragments u. Compute predicted molecular properties http: //www. drugmining. com/

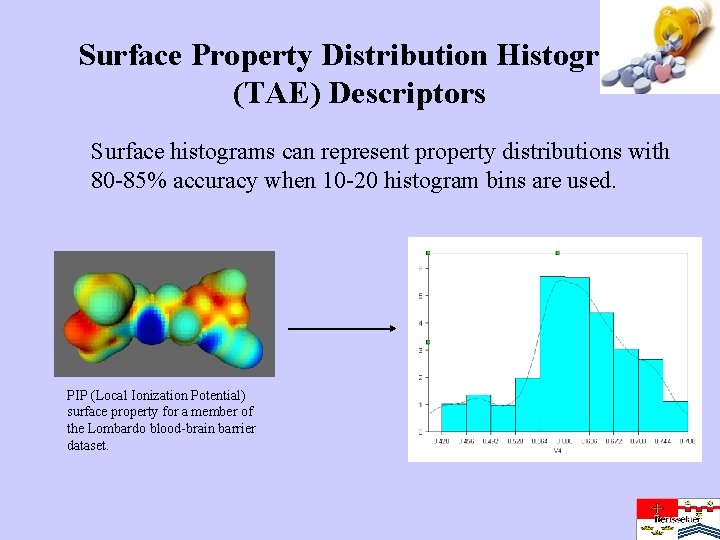
Surface Property Distribution Histogram (TAE) Descriptors Surface histograms can represent property distributions with 80 -85% accuracy when 10 -20 histogram bins are used. PIP (Local Ionization Potential) surface property for a member of the Lombardo blood-brain barrier dataset.

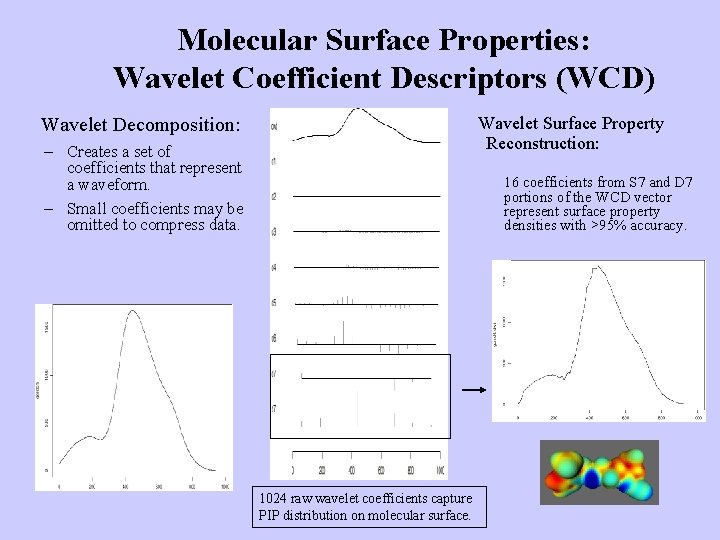
Molecular Surface Properties: Wavelet Coefficient Descriptors (WCD) Wavelet Decomposition: Wavelet Surface Property Reconstruction: – Creates a set of coefficients that represent a waveform. – Small coefficients may be omitted to compress data. 16 coefficients from S 7 and D 7 portions of the WCD vector represent surface property densities with >95% accuracy. 1024 raw wavelet coefficients capture PIP distribution on molecular surface.

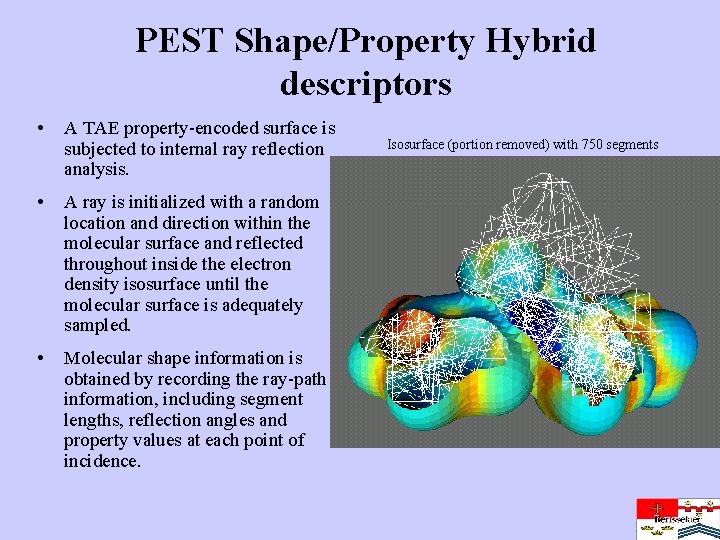
PEST Shape/Property Hybrid descriptors • A TAE property-encoded surface is subjected to internal ray reflection analysis. • A ray is initialized with a random location and direction within the molecular surface and reflected throughout inside the electron density isosurface until the molecular surface is adequately sampled. • Molecular shape information is obtained by recording the ray-path information, including segment lengths, reflection angles and property values at each point of incidence. Isosurface (portion removed) with 750 segments

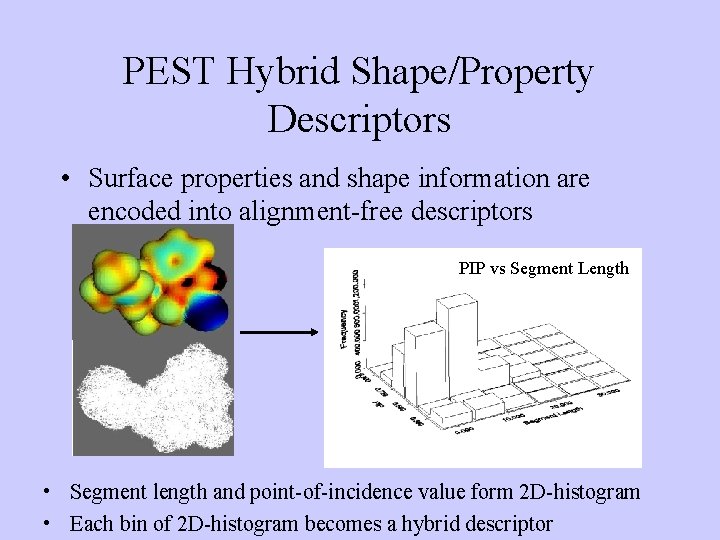
PEST Hybrid Shape/Property Descriptors • Surface properties and shape information are encoded into alignment-free descriptors PIP vs Segment Length • Segment length and point-of-incidence value form 2 D-histogram • Each bin of 2 D-histogram becomes a hybrid descriptor

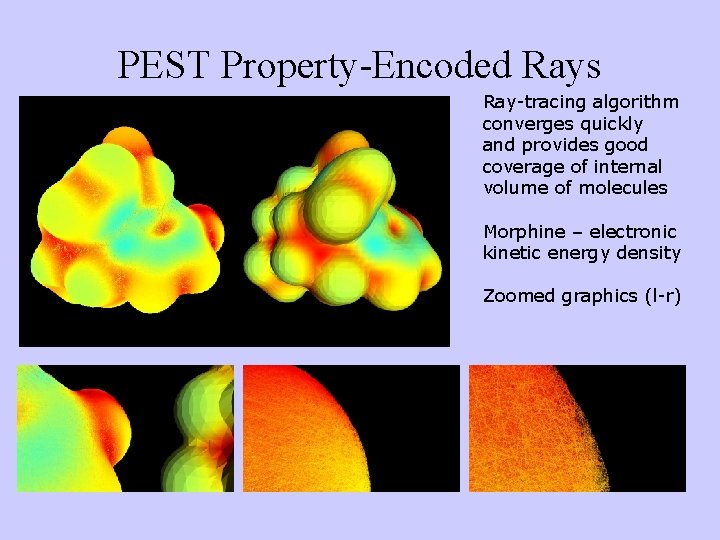
PEST Property-Encoded Rays Ray-tracing algorithm converges quickly and provides good coverage of internal volume of molecules Morphine – electronic kinetic energy density Zoomed graphics (l-r)

PEST Property-Encoded Rays

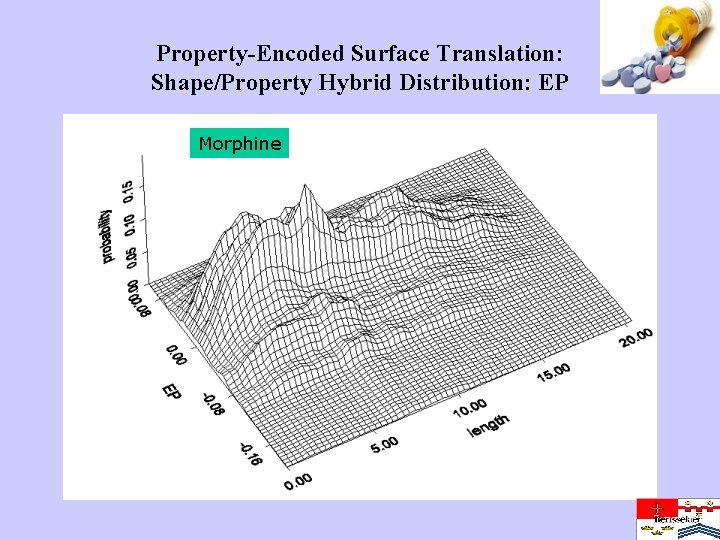
Property-Encoded Surface Translation: Shape/Property Hybrid Distribution: EP Morphine

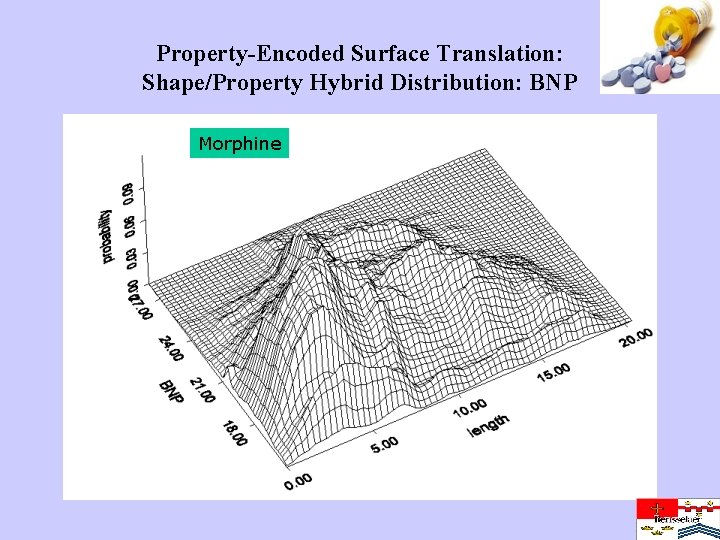
Property-Encoded Surface Translation: Shape/Property Hybrid Distribution: BNP Morphine

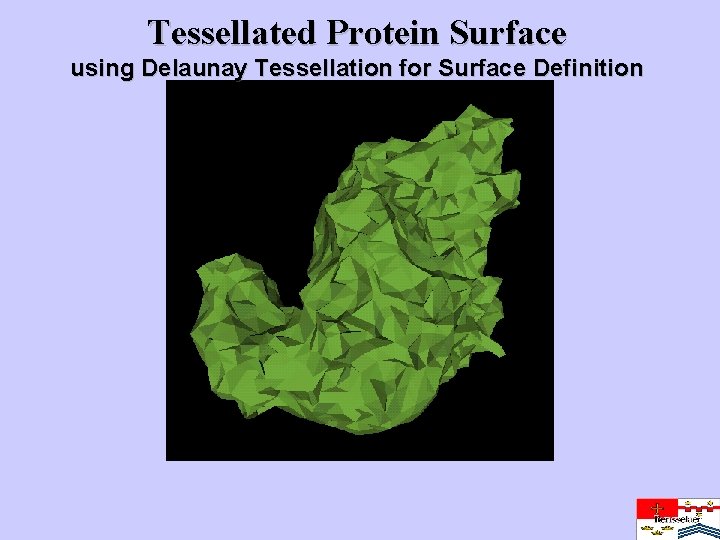
Tessellated Protein Surface using Delaunay Tessellation for Surface Definition

Sliced Surface For 1 A 42 5Å 4Å 6Å

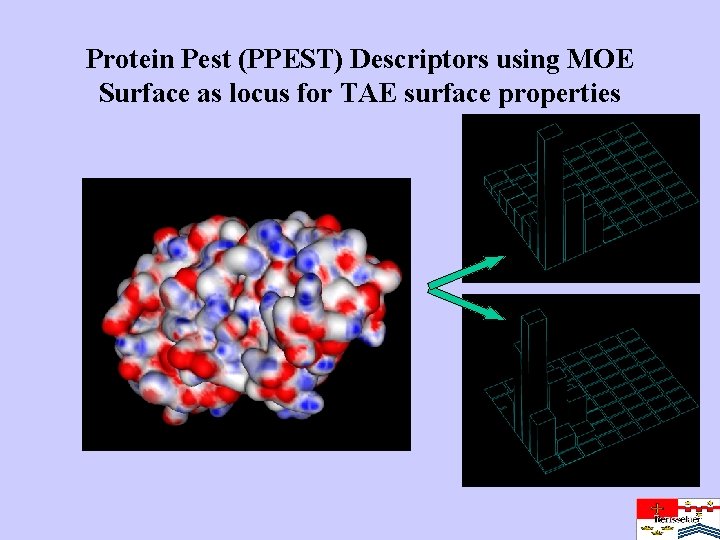
Protein Pest (PPEST) Descriptors using MOE Surface as locus for TAE surface properties

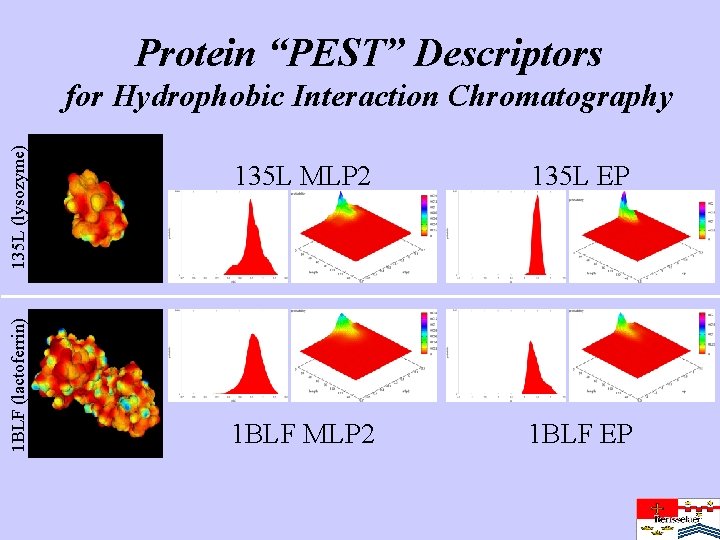
Protein “PEST” Descriptors 1 BLF (lactoferrin) 135 L (lysozyme) for Hydrophobic Interaction Chromatography MLP 2 surface 135 L MLP 2 135 L EP 1 BLF MLP 2 1 BLF EP

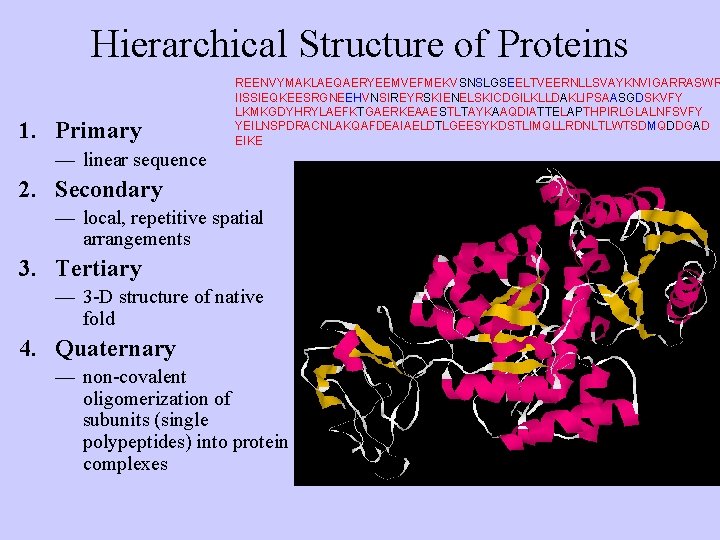
Hierarchical Structure of Proteins 1. Primary REENVYMAKLAEQAERYEEMVEFMEKVSNSLGSEELTVEERNLLSVAYKNVIGARRASWR IISSIEQKEESRGNEEHVNSIREYRSKIENELSKICDGILKLLDAKLIPSAASGDSKVFY LKMKGDYHRYLAEFKTGAERKEAAESTLTAYKAAQDIATTELAPTHPIRLGLALNFSVFY YEILNSPDRACNLAKQAFDEAIAELDTLGEESYKDSTLIMQLLRDNLTLWTSDMQDDGAD EIKE — linear sequence 2. Secondary — local, repetitive spatial arrangements 3. Tertiary — 3 -D structure of native fold 4. Quaternary — non-covalent oligomerization of subunits (single polypeptides) into protein complexes

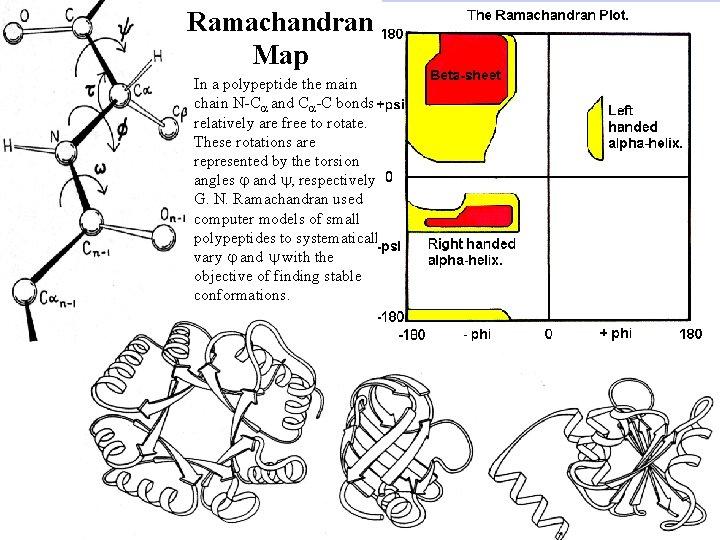
Ramachandran Map In a polypeptide the main chain N-C and C -C bonds relatively are free to rotate. These rotations are represented by the torsion angles and , respectively. G. N. Ramachandran used computer models of small polypeptides to systematically vary and with the objective of finding stable conformations.

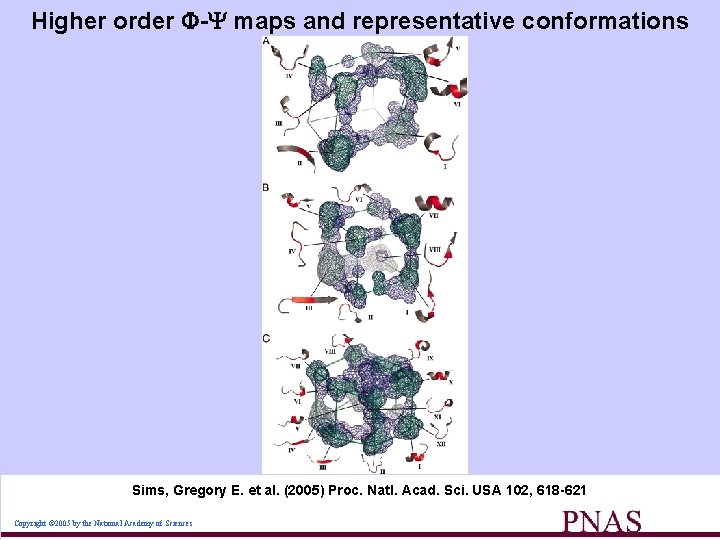
Higher order - maps and representative conformations Sims, Gregory E. et al. (2005) Proc. Natl. Acad. Sci. USA 102, 618 -621 Copyright © 2005 by the National Academy of Sciences
![Protein fingerprint Mihaly Mezei FP 0 ij sign rOirCi rCjrCi FP 1 ij Protein fingerprint — Mihaly Mezei FP 0 ij= sign {[r(Oi)-r(Ci)]. [r(Cj)-r(Ci)]} FP 1 ij=](https://slidetodoc.com/presentation_image_h/e99d84f7752532567d087e571c37c3ef/image-48.jpg)
Protein fingerprint — Mihaly Mezei FP 0 ij= sign {[r(Oi)-r(Ci)]. [r(Cj)-r(Ci)]} FP 1 ij= sign {[r(Ni)-r(Ci)]. [r(Cj)-r(Ci)]}

QSAR assumptions Ø The properties of a chemical are implicit in its — What about effects of the environment? molecular structure All other factors should be held constant in assay; Don’t compare apples to oranges. Ø Molecular structure can be measured and represented with a set of numbers (descriptors or other numerical — But which set of numbers? representation) What descriptors to use? Feature Selection. Ø Compounds with similar structure exhibit similar properties; compounds with dissimilar structure exhibit dissimilar properties — Similar in what way?

Machine Learning Methods “If your experiment needs statistics, you ought to have done a better experiment” - Ernest Rutherford Statistics?