MOLECULAR PHYLOGENETICS AND WHAT TAXONOMY IS THE COURSE

















- Slides: 36

MOLECULAR PHYLOGENETICS AND WHAT TAXONOMY IS THE COURSE ABOUT Ø What is molecular phylogenetics, what is molecular taxonomy Ø Specialities of molecular characters Ø Methods of obtaining the molecular data Ø Methods of data analysis Ø Interpretation of results

PROGRAM FOR THE SEMESTER 5. 10. - Introduction, taxonomy, molecular characters, sekvencing of DNA (Hampl) 12. 10. - Alignment of sequences (Novotný) 19. 10. – Sequence databases and searches in them (Novotný) 26. 10. – Other methods of obtaining of molecular data – multilocus methods (RAPD, RFPL etc. ), microsatelites, minisatelites, SINE elements, protein mass fingerprint (Hampl) 2. 11. - SNP, sequence evolution, calculation of genetic distances (Hampl) 9. 11. – Phylogenetic trees I. - Protein distances, reconstruction of phylogenetic trees from the distance matrix, anatomy of trees (Hampl) 16. 11. - Phylogenetic trees II. Rate heterogeneity, search through the tree space, maximum parsimony (Hampl) 23. 11. - Phylogenetic trees III. - Maximum likelihood, Bayesian metod (Hampl) 30. 11. - Phylogenetic trees IV. - Multigene analyses, robustness of branching, rooting of trees, topology tests, molecular dating (Hampl) 7. 12. – Forensic genetics, DNA barcoding (Hampl) 14. 1. 2020 – Intraspecific phylogeny, population structure and gene flow, phylogeography, examples (Hampl)

EXAM PARTS OF EXAM: Written part (5 examples) – 10 points Oral part (theory) – 8 points ADDITIONAL POINTS Homeworks (earn or loose) – 4 points in total Essay (2 -3 pages) and presentation – 4 points Optional homework – 4 points in total EVALUATION: 11 -13. 5 points – dobře 14 -17. 5 points – velmi dobře above 18 - výborně

MATERIALS FOR STUDY WEB (Hampl): http: //web. natur. cuni. cz/~vlada/moltax/ Moodle: klíč k zápisu „moltax“ BOOKS • Flegr J. Evoluční biologie, Academia 2005. Kapitoly: IX. Evoluce sekvence DNA a XXIV. Molekulární fylogenetika • Avise J. C. Molecular markers, natural history and evolution. Sinauer Associates, Inc. , 2004 • Felsenstein J. Inferring phylogenies. Sinauer Associates, Inc. , 2004 • Lindell Bromham Reading the story of the DNA. Oxford University press 2008. • Higgs P. a Attwood T. K. Bioinformatics and molecular evolution. Blackwell publishing 2005. • Sapp: The new foundation of evolution. Oxford university press 2009 • Yang: Computational Molecular Evolution. Oxford university press 2006 • Hillis a kol. : Molecular Systematics (2 nd edition). Sinauer Associates 1996 • Wiley a Lieberman: Phylogenetics (2 nd edition). Wiley-Blackwell 2011 • An Introduction to Molecular Evolution and Phylogenetics, Lindell Bromham 2016

MOLECULAR TAXONOMY As a discipline Taxonomy (systematics) using molecular characters Taxonomy (systematics) It tries to catalog biodiversity and organize it into a system of usually hierarchically arranged groups. Difference in the sequence of DNA (and proteins). It does not include features on other molecules (lipids, polysaccharides, proteoglycans, tertiary protein structures, etc. )

(MOLECULAR) PHYLOGENETICS According to the majority opinion of taxonomists, the best natural system of organisms is the one that reflects the course of their phylogeny. Phylogenetics - deals with reconstruction of the origin and development of lineages of organisms. It reconstructs the course of cladogenesis (branching), but also focuses on the anagenesis the changes of character states of organisms within the line. Tree of life (TOL) = objectively existing entity Phylogenetic reconstructions = hypotheses about TOL Organismal taxa are historical groups (kins)

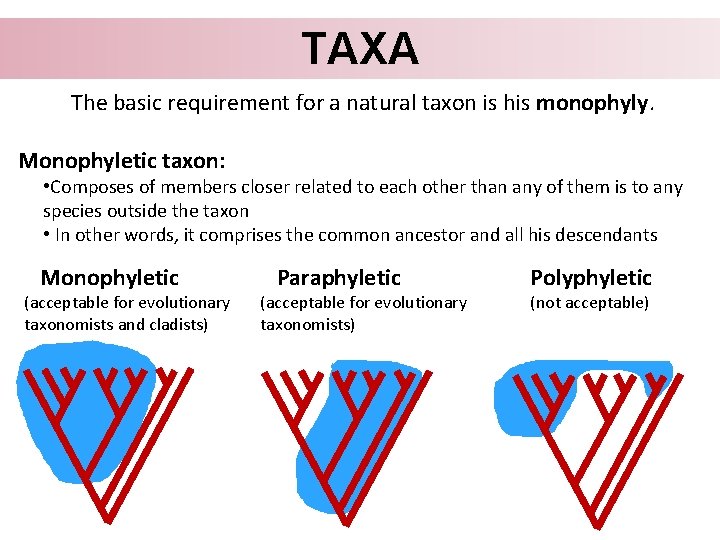
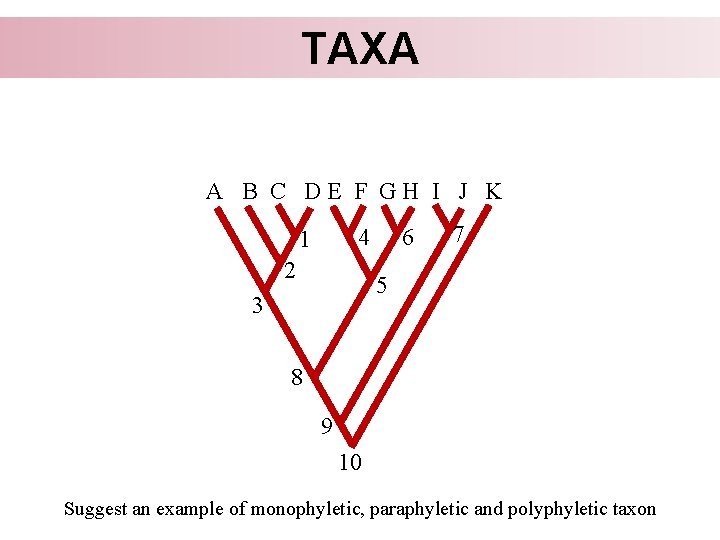
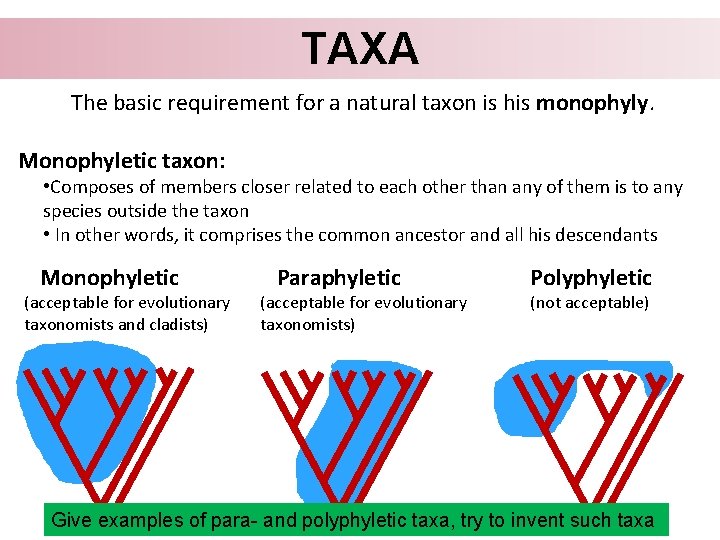
TAXA The basic requirement for a natural taxon is his monophyly. Monophyletic taxon: • Composes of members closer related to each other than any of them is to any species outside the taxon • In other words, it comprises the common ancestor and all his descendants Monophyletic (acceptable for evolutionary taxonomists and cladists) Paraphyletic (acceptable for evolutionary taxonomists) Polyphyletic (not acceptable)

TAXA A B C DE F GH I J K 6 4 1 2 7 5 3 8 9 10 Suggest an example of monophyletic, paraphyletic and polyphyletic taxon

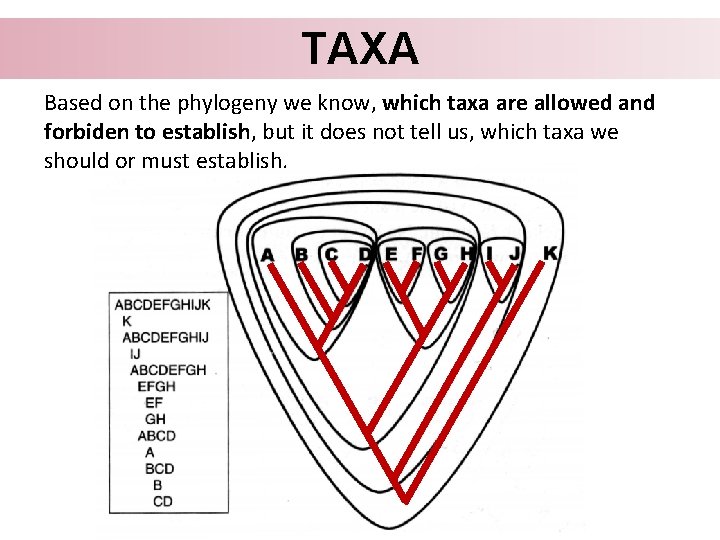
TAXA Based on the phylogeny we know, which taxa are allowed and forbiden to establish, but it does not tell us, which taxa we should or must establish.

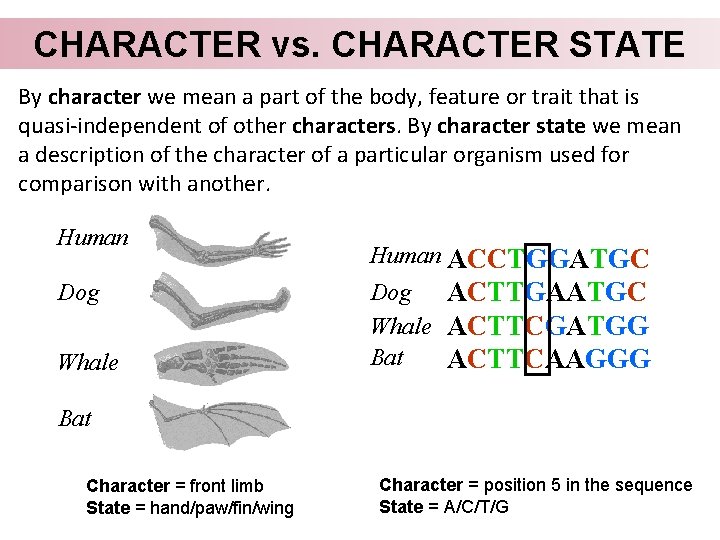
CHARACTER vs. CHARACTER STATE By character we mean a part of the body, feature or trait that is quasi-independent of other characters. By character state we mean a description of the character of a particular organism used for comparison with another. Human Dog Whale Human ACCTGGATGC Dog ACTTGAATGC Whale ACTTCGATGG Bat ACTTCAAGGG Bat Character = front limb State = hand/paw/fin/wing Character = position 5 in the sequence State = A/C/T/G

HOMOLOGY Homologies are similarities between complex structures or patterns that are caused by the continuity of biological information. (Riedl and Hazsprunar) We must realize whether we are considering the homology of a character or a character state. It is important to compare states of the homologous characters How do we recognise homologous characters?

https: //whiteboard. fi/c 2 t 9 f

HOMOLOGY OF CHARACTER SUPPORTS Homology is indicated by: • Similar position ACCTGGATGC ACTTGAATGC ACTTCGATGG ACTTCAAGGG Sequence alignment • Similarity in details

HOMOLOGY OF CHARACTER REJECTS Homology of character is rejected by presence of two states in one organism (test of conjunction). This situation indicates that we are not comparing states of the same character.

OPTIONAL ASSIGNMENT (1 POINT) Different body segments may be homologous at certain level Write a short thought on this elaborating: • the rationale for our belief that there is such homology • how does it fit to the definition of homology by Riedl and Hazsprunar • find equivalent in the molecular data Deadline 26. 10. 9: 00

HOMOLOGY OF CHARACTER STATES Homology of two character states is indicates by similarity (auxiliary principle) Homology of two character states is rejected by incongruence with distribution of character states of other character. Mutliple sequence alignment (MSA)

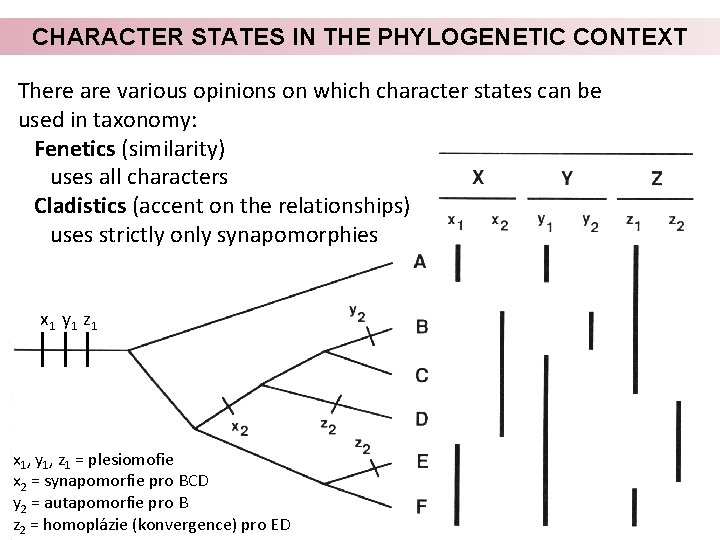
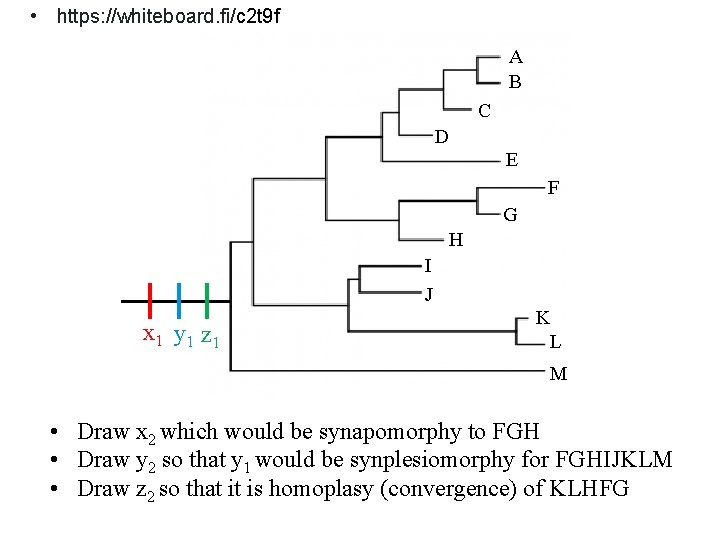
CHARACTER STATES IN THE PHYLOGENETIC CONTEXT There are various opinions on which character states can be used in taxonomy: Fenetics (similarity) uses all characters Cladistics (accent on the relationships) uses strictly only synapomorphies x 1 y 1 z 1 x 1, y 1, z 1 = plesiomofie x 2 = synapomorfie pro BCD y 2 = autapomorfie pro B z 2 = homoplázie (konvergence) pro ED

• https: //whiteboard. fi/c 2 t 9 f A B C D E F G H I J x 1 y 1 z 1 K L M • Draw x 2 which would be synapomorphy to FGH • Draw y 2 so that y 1 would be synplesiomorphy for FGHIJKLM • Draw z 2 so that it is homoplasy (convergence) of KLHFG

TAXA The basic requirement for a natural taxon is his monophyly. Monophyletic taxon: • Composes of members closer related to each other than any of them is to any species outside the taxon • In other words, it comprises the common ancestor and all his descendants Monophyletic (acceptable for evolutionary taxonomists and cladists) Paraphyletic (acceptable for evolutionary taxonomists) Polyphyletic (not acceptable) Give examples of para- and polyphyletic taxa, try to invent such taxa

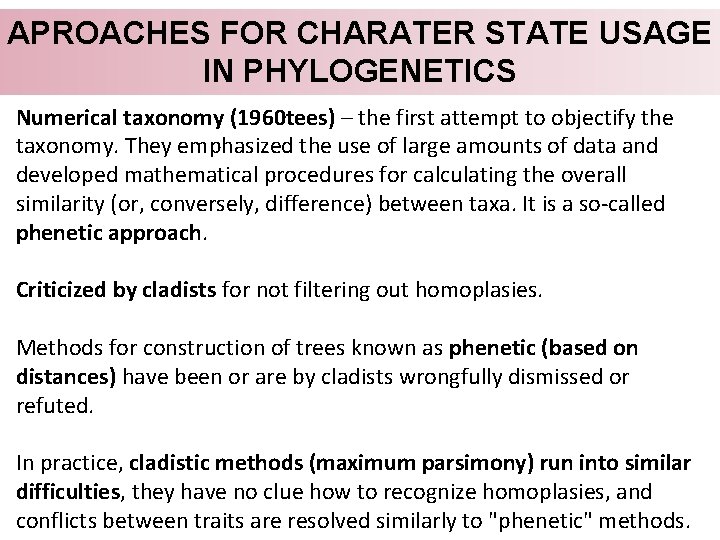
APROACHES FOR CHARATER STATE USAGE IN PHYLOGENETICS Numerical taxonomy (1960 tees) – the first attempt to objectify the taxonomy. They emphasized the use of large amounts of data and developed mathematical procedures for calculating the overall similarity (or, conversely, difference) between taxa. It is a so-called phenetic approach. Criticized by cladists for not filtering out homoplasies. Methods for construction of trees known as phenetic (based on distances) have been or are by cladists wrongfully dismissed or refuted. In practice, cladistic methods (maximum parsimony) run into similar difficulties, they have no clue how to recognize homoplasies, and conflicts between traits are resolved similarly to "phenetic" methods.

MOLECULAR TAXONOMY As a discipline Taxonomy (systematics) using molecular characters Taxonomy (systematics) It tries to catalog biodiversity and organize it into a system of usually hierarchically arranged groups. Difference in the sequnce of DNA (proteins). It does not include features on other molecules (lipids, polysaccharides, proteoglycans, tertiary protein structures, etc. )

SURPRISINGLY HIGH AMOUNT OF POLYMORPHISM Differ in 3 millions of nucleotides on average If these different alleles of genes affected fitness, they would be quickly eliminated or fixed by natural selection, and no polymorphism would be observed in the given positions.

NEUTRAL THEORY OF MOLECULAR EVOLUTION Differ in 3 millions of nucleotides on average The vast majority of substitutions at the DNA level are selectively neutral, mutants have the same fitness. These mutations are invisible for selection and their fixation or elimination causes genetic drift. It is slow in large populations, both alleles persist there for a long time and we detect them as polymorphisms.

GENETIC DRIFT

NEUTRAL THEORY OF MOLECULAR EVOLUTION NEUTRÁLNÍ TEORIE EVOLUCE To make things clear: • Neutral theory does not claim that most genes are dispensable, but claims that most forms (alleles) of the same gene are functionally equally good. • Neutral theory does not claim that there are no mutations with a deleterious effect that are eliminated by natural selection, but claims that there a minority of such mutations. • Neutral theory does not reject Darwinist adaptive evolution driven by natural selection, but argues that most mutations are "invisible" to natural selection and do not contribute to adaptive evolution. Neutral theory best explains where so many polymorphisms (differences) in DNA come from.

ADVANTAGES OF MOLECULAR CHARACTERS 1. Are genetic We know how they are inherited, it does not depend on the environment or the genetic background. This is exactly the level where evolutionary innovations arise - mutations in DNA. 2. There a huge number of them: The size of genomes ranges from 0. 5 * 106 - 600 * 109. The human genome contains over 3 billion base pairs. It is estimated that people differ from each other in 0. 1%, i. e. 3 million bases.

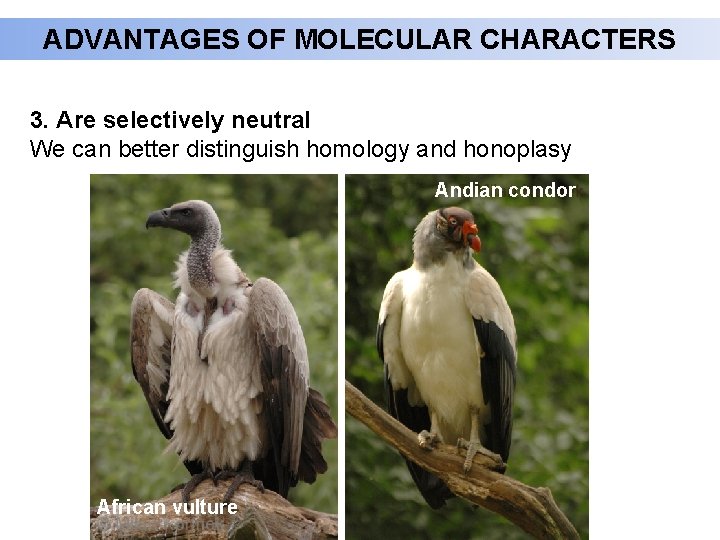
ADVANTAGES OF MOLECULAR CHARACTERS 3. Are selectively neutral We can better distinguish homology and honoplasy Andian Supcondor africký African vulture

ADVANTAGES OF MOLECULAR CHARACTERS 4. Are useful for the most distant comparisons… ACCTGGATGC ACTTGAATGC ACTTCGATGG ACTTCAAGGG

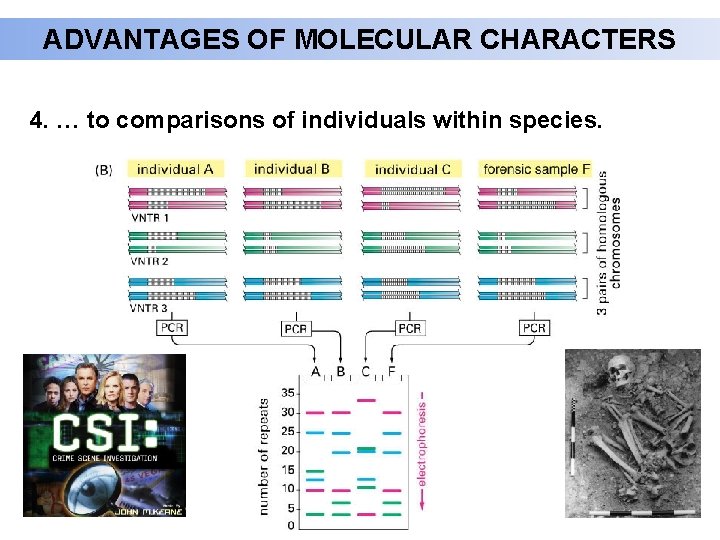
ADVANTAGES OF MOLECULAR CHARACTERS 4. … to comparisons of individuals within species.

ADVANTAGES OF MOLECULAR CHARACTERS 5. Can be simply described 6. Are independent ACCTGGATGC ACTTGAATGC ACTTCGATGG ACTTCAAGGG 2 1 7. Easily weighted 8. The level of uncertainity can be evaluated more easily

ADVANTAGES OF MOLECULAR CHARACTERS • Molecular clock • Informative about the population history

DISADVANTAGES OF MOLECULAR CHARACTERS • No information about anagenesis • Price • Destructive methods (sometimes)

HISTORY – 60’s Linus Pauling, Emile Zuckerkandl (Molecules as Documents of Evolutionary History, 1964) Robert Sokal, Peter Sneath (Numerical taxonomy, 1963) Willi Henning (Phylogenetic systematics, 1966) Luigi Cavalli-Sforza, Anthony Edwards (maximum parsimony and maximum likelihood, 1963 -1966)

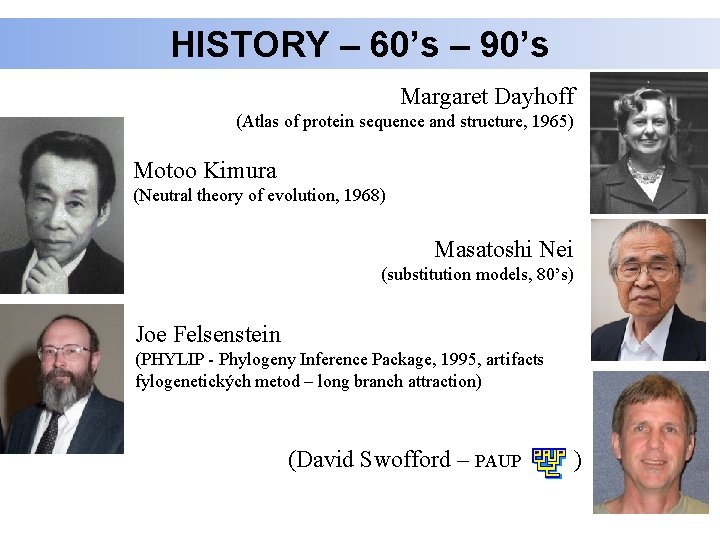
HISTORY – 60’s – 90’s Margaret Dayhoff (Atlas of protein sequence and structure, 1965) Motoo Kimura (Neutral theory of evolution, 1968) Masatoshi Nei (substitution models, 80’s) Joe Felsenstein (PHYLIP - Phylogeny Inference Package, 1995, artifacts fylogenetických metod – long branch attraction) (David Swofford – PAUP )

SUMMARY • The natural classification is based on knowledge of the phylogeny of organisms • We must compare homologous characters • It is allowed to create only monophyletic or paraphyletic taxa • Molecular characters have many important advantages • Molecular features are suitable for studying the study of cladogenesis, not anagenesis • Molecular traits are formed mainly by neutral evolution and genetic drift contributes to their fixation

DNA SEQUENCING Self study assignment – Sequencing technologies Deadline 26. 10. 9: 00