MOLECULAR PATHOLOGY IN MELANOMA Dr Jos Luis Rodrguez









- Slides: 39

MOLECULAR PATHOLOGY IN MELANOMA Dr José Luis Rodríguez Peralto Hospital Universitario 12 de Octubre, Madrid, Spain

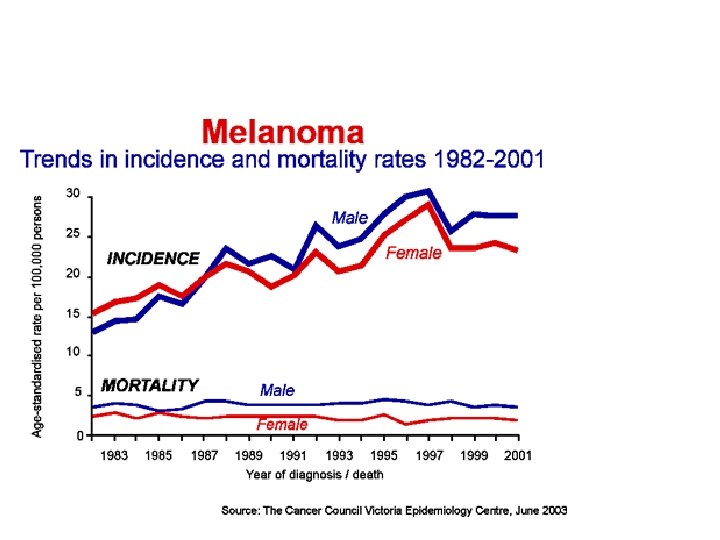
AN IMPORTANT HEALTH PROBLEM • Melanoma is one of the most aggressive tumours (79% of skin cancer death) • In Europe, 2. 5% of all cancers. 1 -2% death by cancer (5000 deaths by melanoma a year in Europe) • Incidence has increased dramatically in last decades (15 times in last 50 years) • Incidence in Spain: 15: 100. 000 inhabitants (depend on regions) • The rate of mortality has not increased in the same average as incidence (early diagnostic) • Extremely bad prognosis, especially in advanced stages. Resistance to chemotherapy


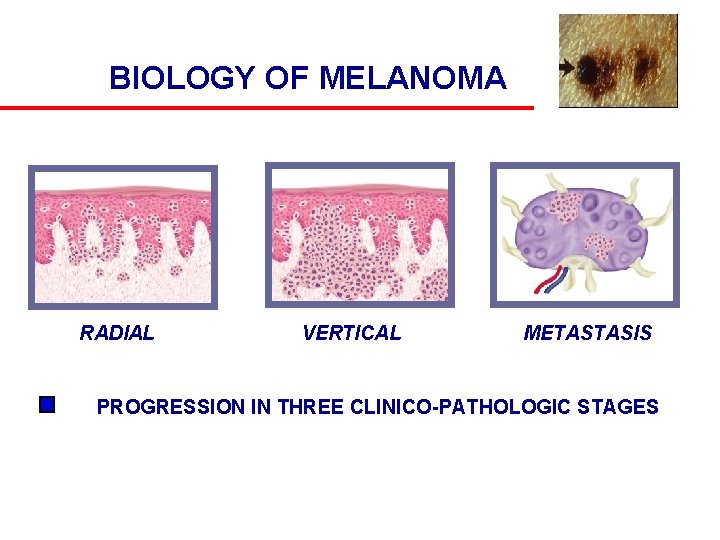
BIOLOGY OF MELANOMA RADIAL VERTICAL METASTASIS PROGRESSION IN THREE CLINICO-PATHOLOGIC STAGES

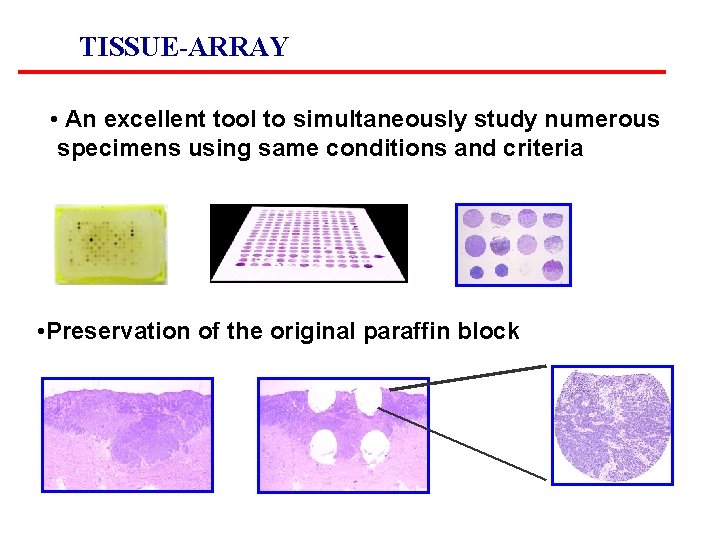
TISSUE-ARRAY • An excellent tool to simultaneously study numerous specimens using same conditions and criteria • Preservation of the original paraffin block

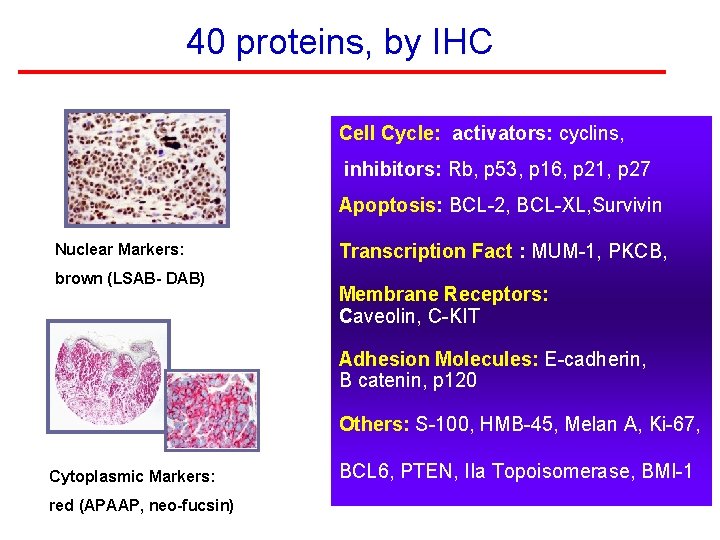
40 proteins, by IHC Cell Cycle: activators: cyclins, inhibitors: Rb, p 53, p 16, p 21, p 27 Apoptosis: BCL-2, BCL-XL, Survivin Nuclear Markers: brown (LSAB- DAB) Transcription Fact : MUM-1, PKCB, Membrane Receptors: Caveolin, C-KIT Adhesion Molecules: E-cadherin, B catenin, p 120 Others: S-100, HMB-45, Melan A, Ki-67, Cytoplasmic Markers: red (APAAP, neo-fucsin) BCL 6, PTEN, IIa Topoisomerase, BMI-1

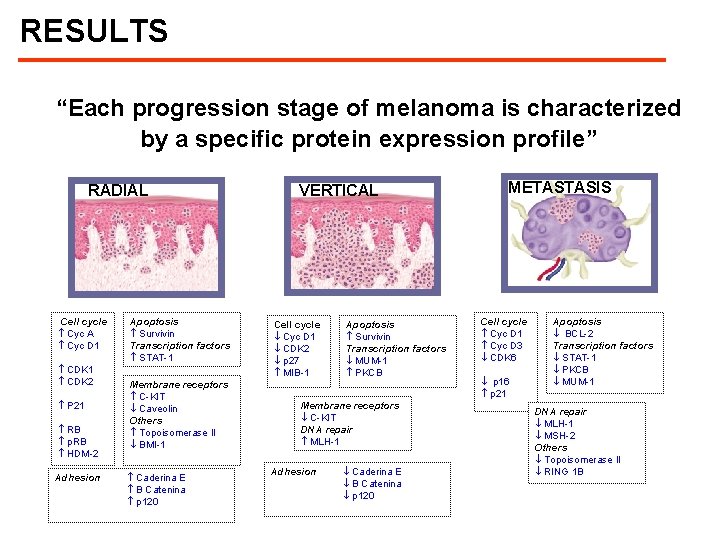
RESULTS “Each progression stage of melanoma is characterized by a specific protein expression profile” RADIAL Cell cycle Cyc A Cyc D 1 CDK 2 P 21 RB p. RB HDM-2 Adhesion Apoptosis Survivin Transcription factors STAT-1 Membrane receptors C-KIT Caveolin Others Topoisomerase II BMI-1 Caderina E B Catenina p 120 VERTICAL Cell cycle Cyc D 1 CDK 2 p 27 MIB-1 Apoptosis Survivin Transcription factors MUM-1 PKCB Membrane receptors C-KIT DNA repair MLH-1 Adhesion Caderina E B Catenina p 120 METASTASIS Cell cycle Cyc D 1 Cyc D 3 CDK 6 p 16 p 21 Apoptosis BCL-2 Transcription factors STAT-1 PKCB MUM-1 DNA repair MLH-1 MSH-2 Others Topoisomerase II RING 1 B

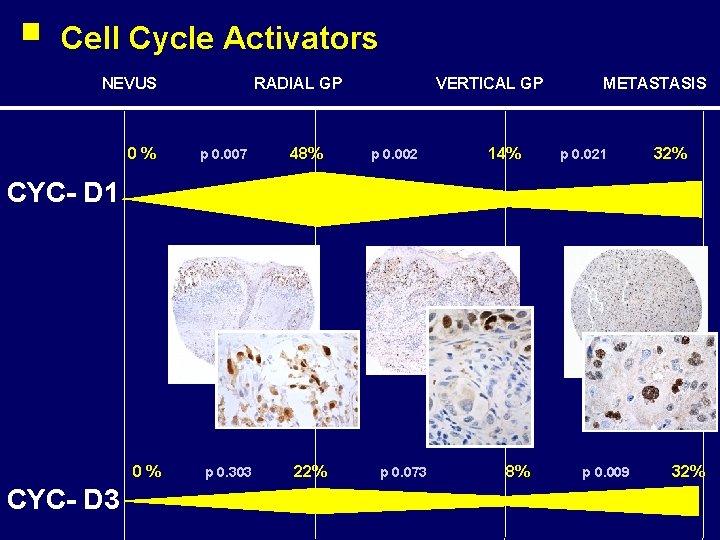
Cell Cycle Activators NEVUS 0% RADIAL GP p 0. 007 48% VERTICAL GP p 0. 002 14% METASTASIS p 0. 021 32% CYC- D 1 0% CYC- D 3 p 0. 303 22% p 0. 073 8% p 0. 009 32%

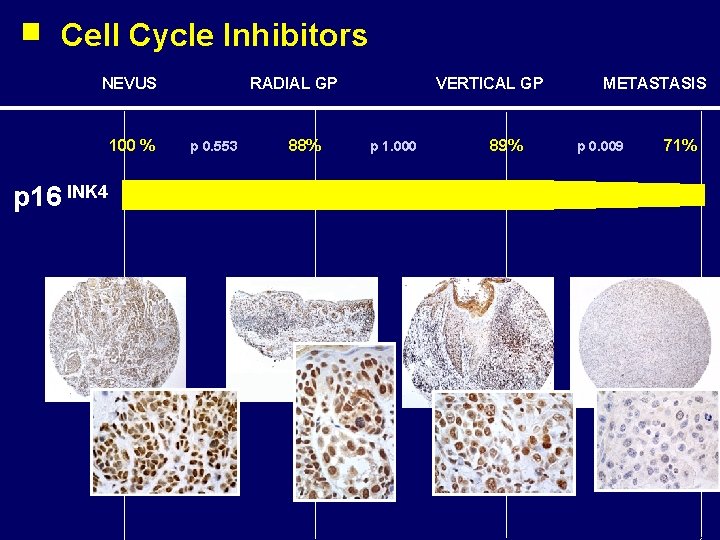
Cell Cycle Inhibitors NEVUS 100 % p 16 INK 4 RADIAL GP p 0. 553 88% VERTICAL GP p 1. 000 89% METASTASIS p 0. 009 71%

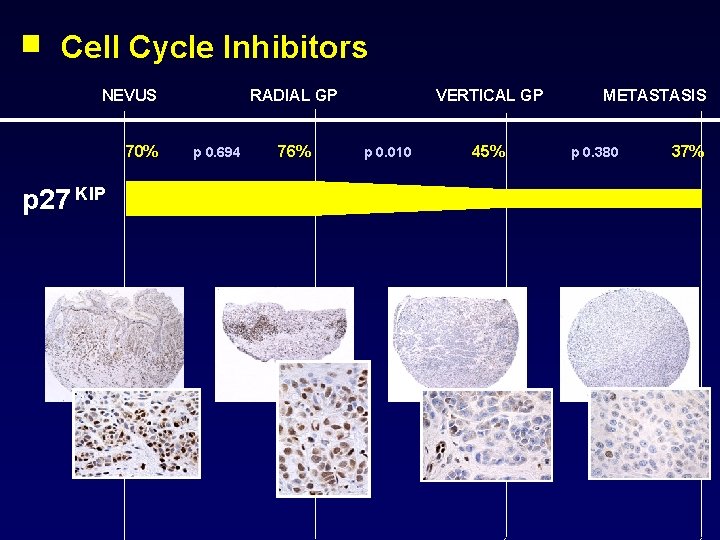
Cell Cycle Inhibitors NEVUS 70% p 27 KIP RADIAL GP p 0. 694 76% VERTICAL GP p 0. 010 45% METASTASIS p 0. 380 37%

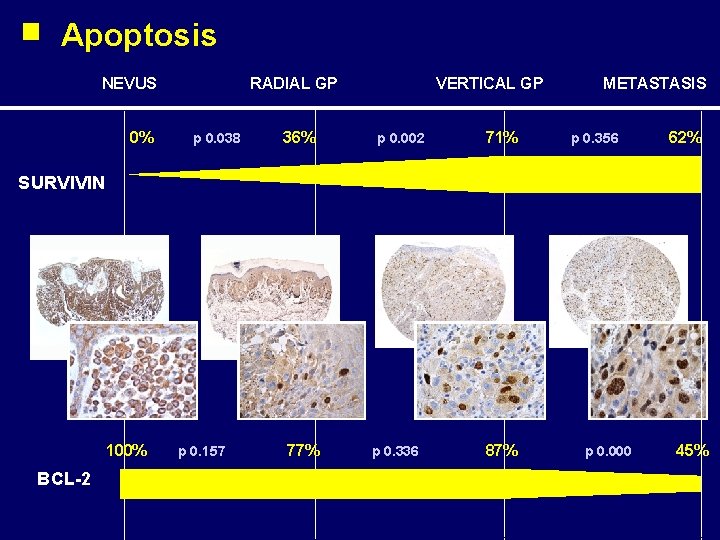
Apoptosis NEVUS 0% RADIAL GP p 0. 038 36% VERTICAL GP p 0. 002 71% METASTASIS p 0. 356 62% SURVIVIN 100% BCL-2 p 0. 157 77% p 0. 336 87% p 0. 000 45%

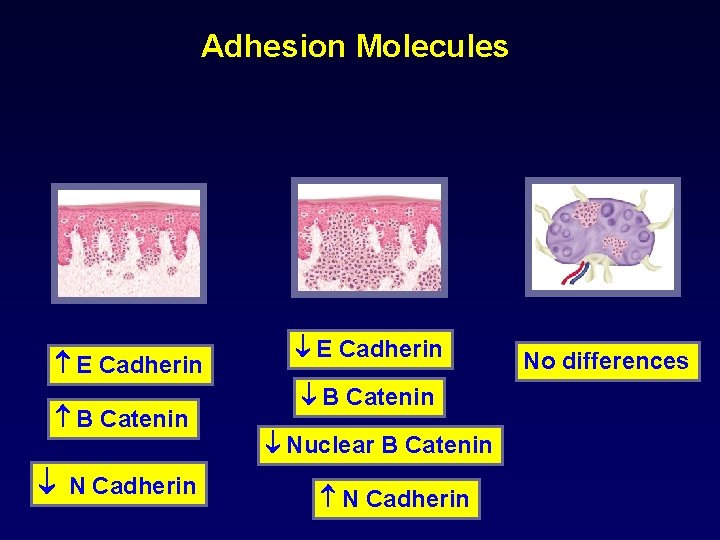
Adhesion Molecules E Cadherin B Catenin N Cadherin E Cadherin B Catenin Nuclear B Catenin N Cadherin No differences

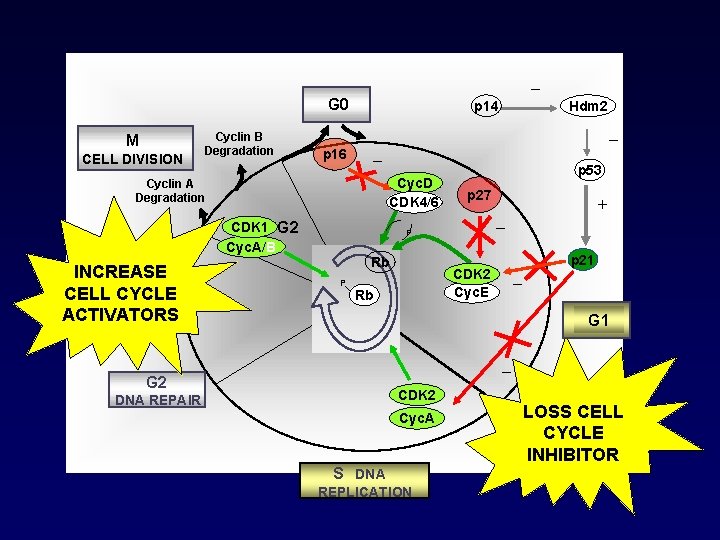
_ G 0 M CELL DIVISION Cyclin B Degradation p 14 _ p 16 _ Cyc. D CDK 4/6 Cyclin A Degradation CDK 1 G 2 Cyc. A/B INCREASE CELL CYCLE ACTIVATORS Hdm 2 p 53 p 27 _ P Rb P _ CDK 2 Cyc. E Rb + p 21 G 1 _ G 2 CDK 2 DNA REPAIR Cyc. A S DNA REPLICATION LOSS CELL CYCLE INHIBITOR

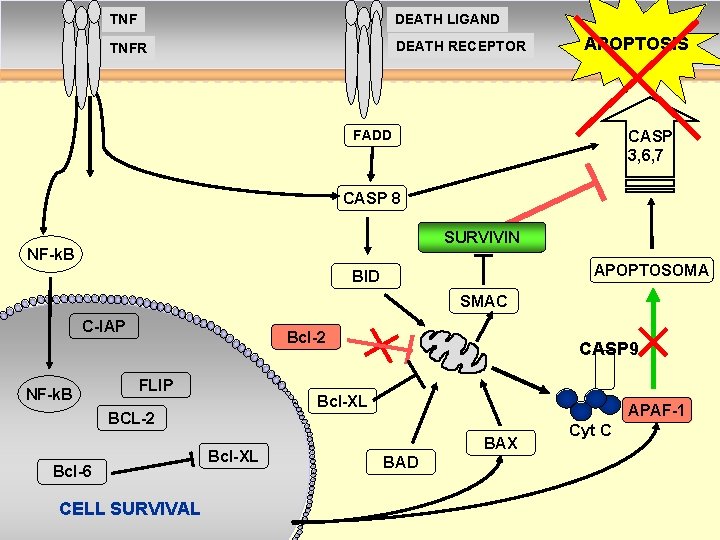
TNF DEATH LIGAND TNFR DEATH RECEPTOR APOPTOSIS CASP 3, 6, 7 FADD CASP 8 SURVIVIN NF-k. B APOPTOSOMA BID SMAC C-IAP Bcl-2 Surv NF-k. B FLIP Bcl-XL BCL-2 Bcl-6 CELL SURVIVAL CASP 9 Bcl-XL p 53 APAF-1 BAX BAD Cyt C

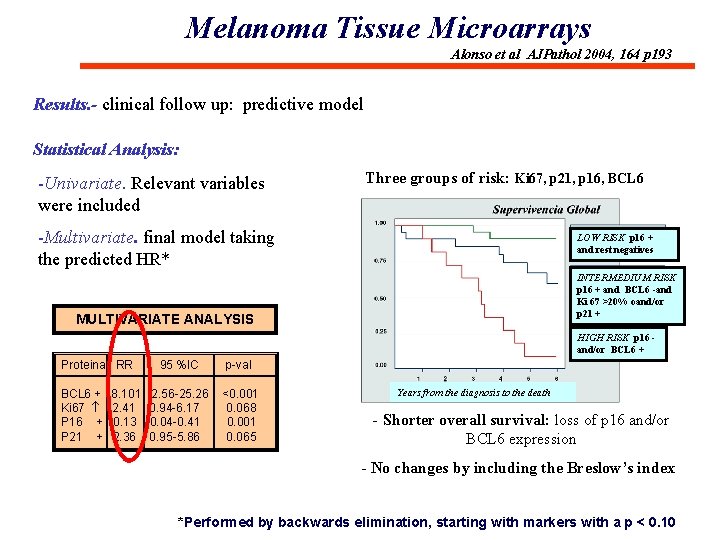
Melanoma Tissue Microarrays Alonso et al AJPathol 2004, 164 p 193 Results. - clinical follow up: predictive model Statistical Analysis: -Univariate. Relevant variables were included Three groups of risk: Ki 67, p 21, p 16, BCL 6 -Multivariate. final model taking the predicted HR* LOW RISK p 16 + and rest negatives INTERMEDIUM RISK p 16 + and BCL 6 -and Ki 67 >20% oand/or p 21 + MULTIVARIATE ANALYSIS HIGH RISK p 16 and/or BCL 6 + Proteina RR BCL 6 + Ki 67 P 16 + P 21 + 8. 101 2. 41 0. 13 2. 36 95 %IC p-val 2. 56 -25. 26 0. 94 -6. 17 0. 04 -0. 41 0. 95 -5. 86 <0. 001 0. 068 0. 001 0. 065 Years from the diagnosis to the death - Shorter overall survival: loss of p 16 and/or BCL 6 expression - No changes by including the Breslow’s index *Performed by backwards elimination, starting with markers with a p < 0. 10

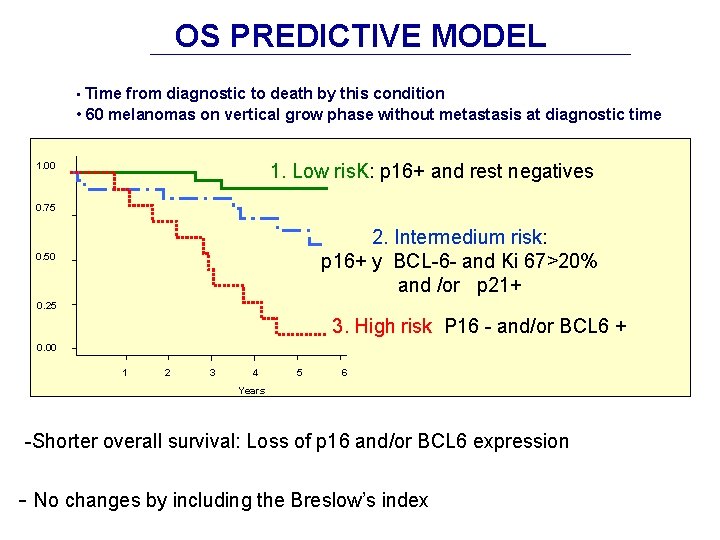
OS PREDICTIVE MODEL • Time from diagnostic to death by this condition • 60 melanomas on vertical grow phase without metastasis at diagnostic time 1. 00 1. Low ris. K: p 16+ and rest negatives 0. 75 2. Intermedium risk: p 16+ y BCL-6 - and Ki 67>20% and /or p 21+ 0. 50 0. 25 3. High risk P 16 - and/or BCL 6 + 0. 00 1 2 3 4 5 6 Years -Shorter overall survival: Loss of p 16 and/or BCL 6 expression - No changes by including the Breslow’s index


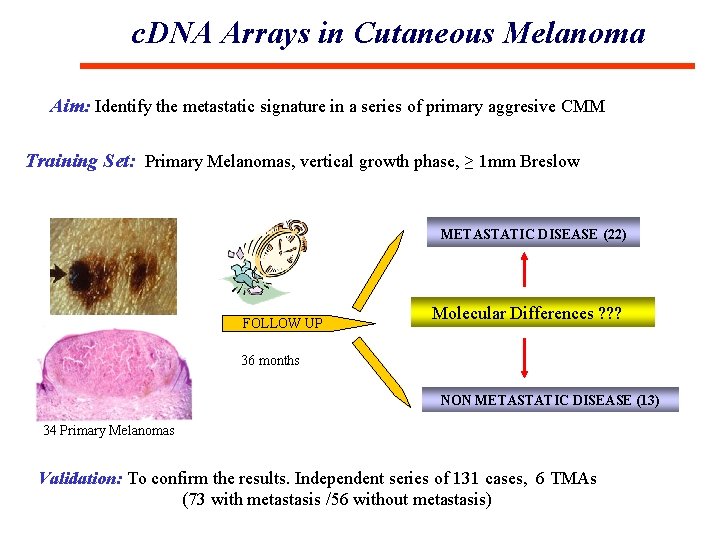
c. DNA Arrays in Cutaneous Melanoma Aim: Identify the metastatic signature in a series of primary aggresive CMM Training Set: Primary Melanomas, vertical growth phase, ≥ 1 mm Breslow METASTATIC DISEASE (22) FOLLOW UP Molecular Differences ? ? ? 36 months NON METASTATIC DISEASE (13) 34 Primary Melanomas Validation: To confirm the results. Independent series of 131 cases, 6 TMAs (73 with metastasis /56 without metastasis)

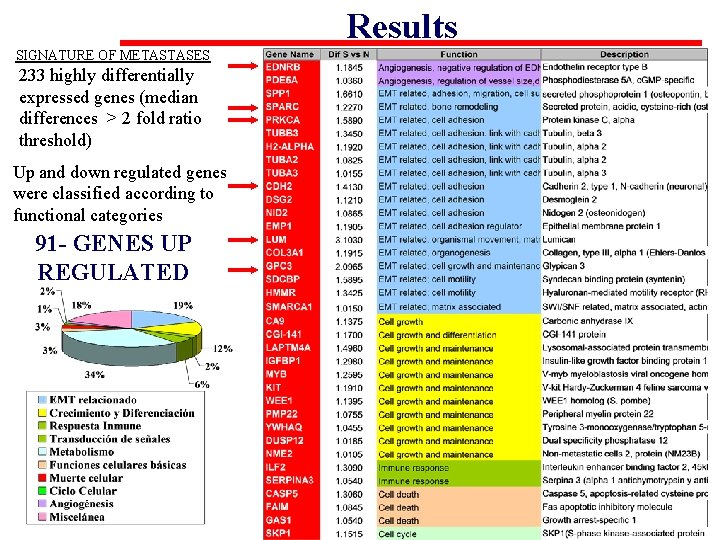
Results SIGNATURE OF METASTASES 233 highly differentially expressed genes (median differences > 2 fold ratio threshold) Up and down regulated genes were classified according to functional categories 91 - GENES UP REGULATED

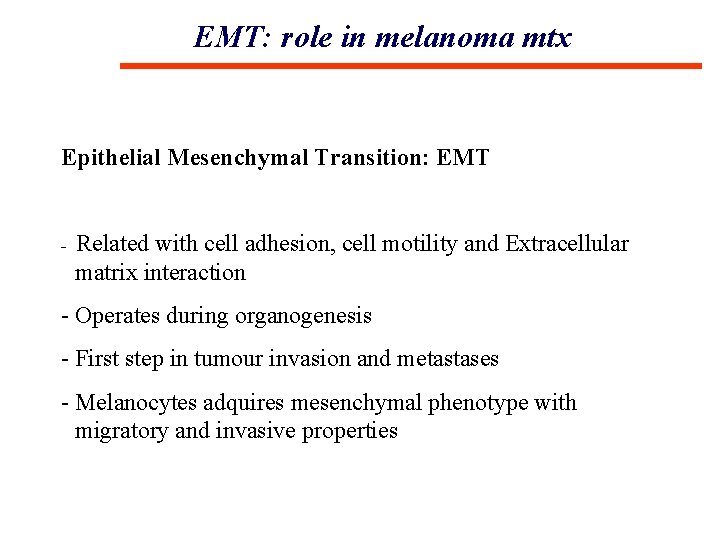
EMT: role in melanoma mtx Epithelial Mesenchymal Transition: EMT - Related with cell adhesion, cell motility and Extracellular matrix interaction - Operates during organogenesis - First step in tumour invasion and metastases - Melanocytes adquires mesenchymal phenotype with migratory and invasive properties

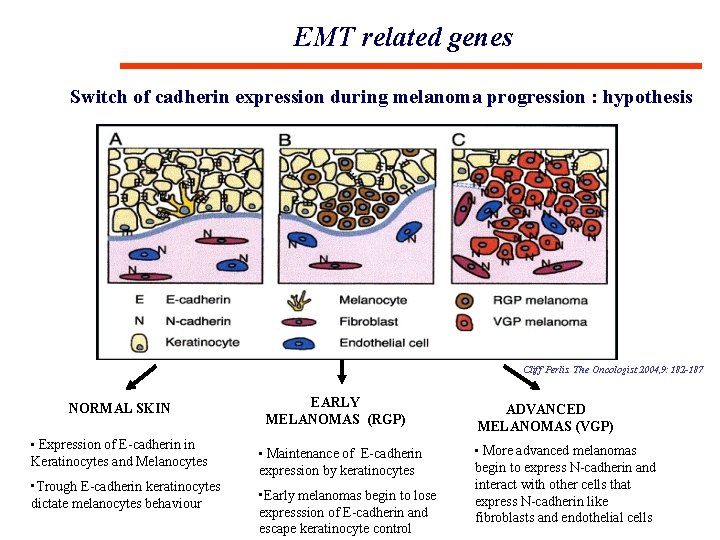
EMT related genes Switch of cadherin expression during melanoma progression : hypothesis Cliff Perlis. The Oncologist 2004, 9: 182 -187 NORMAL SKIN • Expression of E-cadherin in Keratinocytes and Melanocytes • Trough E-cadherin keratinocytes dictate melanocytes behaviour EARLY MELANOMAS (RGP) • Maintenance of E-cadherin expression by keratinocytes • Early melanomas begin to lose expresssion of E-cadherin and escape keratinocyte control ADVANCED MELANOMAS (VGP) • More advanced melanomas begin to express N-cadherin and interact with other cells that express N-cadherin like fibroblasts and endothelial cells

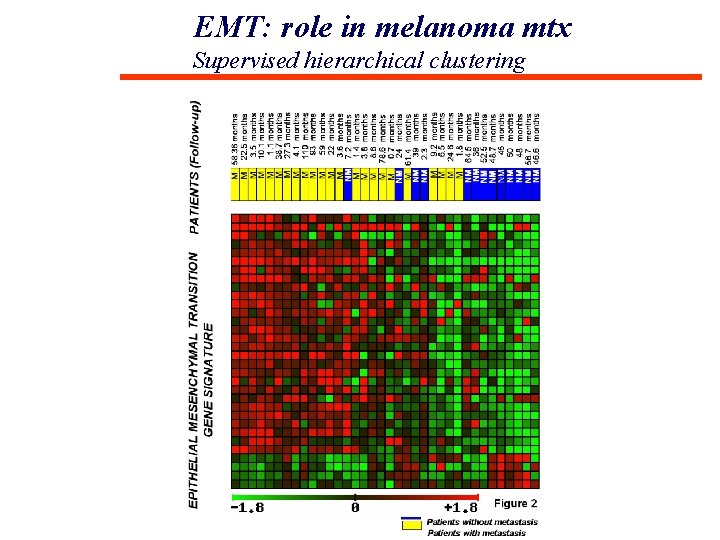
EMT: role in melanoma mtx Supervised hierarchical clustering

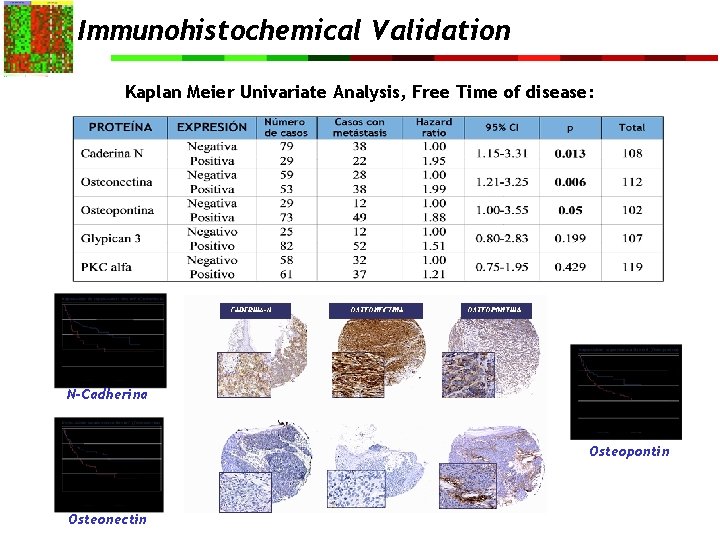
Immunohistochemical Validation Kaplan Meier Univariate Analysis, Free Time of disease: N-Cadherina Osteopontin Osteonectin

A high-throughput study in melanoma identifies Epithelial-Mesenchymal Transition as a major determinant of metastasis Soledad R. Alonso 1, Lorraine Tracey 1, Pablo Ortiz 4, Beatriz Pérez-Gómez 5, José Palacios 1, Marina Pollán 5, Juan Linares 6, Salvio Serrano 7, Ana I. Sáez-Castillo 6, Lydia Sánchez 2, Raquel Pajares 2, Abel Sánchez-Aguilera 1 , Miguel A. Piris 1 and José L. Rodríguez-Peralto 3. From the Molecular Pathology Programme 1 and Histology and Immunohistochemistry Unit 2, Centro Nacional de Investigaciones Oncológicas (CNIO), Madrid; Departments of Pathology 3 and Dermatology 4, Hospital Universitario 12 de Octubre, Madrid; Centro Nacional de Epidemiología, Instituto de Salud Carlos III 5, Madrid; Departments of Pathology 6 and Dermatology 7, Hospital Universitario San Cecilio, Granada, Spain. Cancer Research 2007 67: 3450 -3460

MOLECULAR PATHOLOGY IN MELANOMA TREATMENT

TREATMENT • Surgery: Surgical excision with free margin. • Elective lymph node dissection • Surgical excision of isolated metastasis • Regional Radiotherapy • Pharmacological Immunomodulator • Chemotherapy…

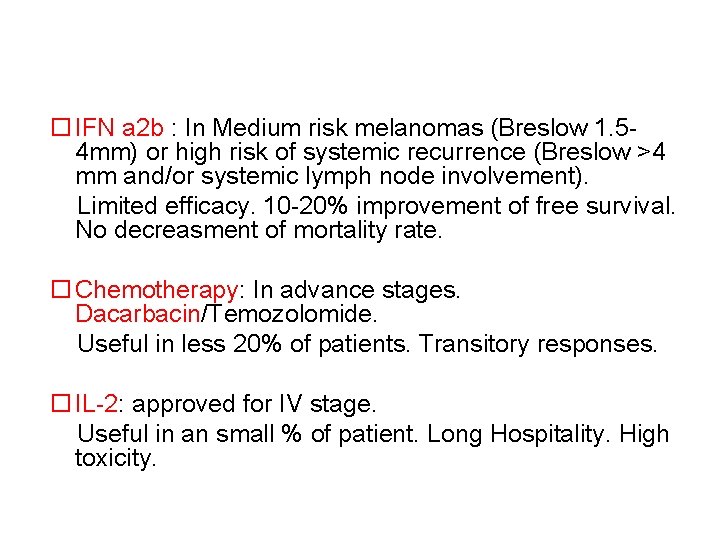
IFN a 2 b : In Medium risk melanomas (Breslow 1. 54 mm) or high risk of systemic recurrence (Breslow >4 mm and/or systemic lymph node involvement). Limited efficacy. 10 -20% improvement of free survival. No decreasment of mortality rate. Chemotherapy: In advance stages. Dacarbacin/Temozolomide. Useful in less 20% of patients. Transitory responses. IL-2: approved for IV stage. Useful in an small % of patient. Long Hospitality. High toxicity.

… None systemic treatment has demonstrated a significant survival in mestastatic melanoma…

. . . “During 30 years, there was no significant advances in treatment of melanoma”…

… ¿A new era? …

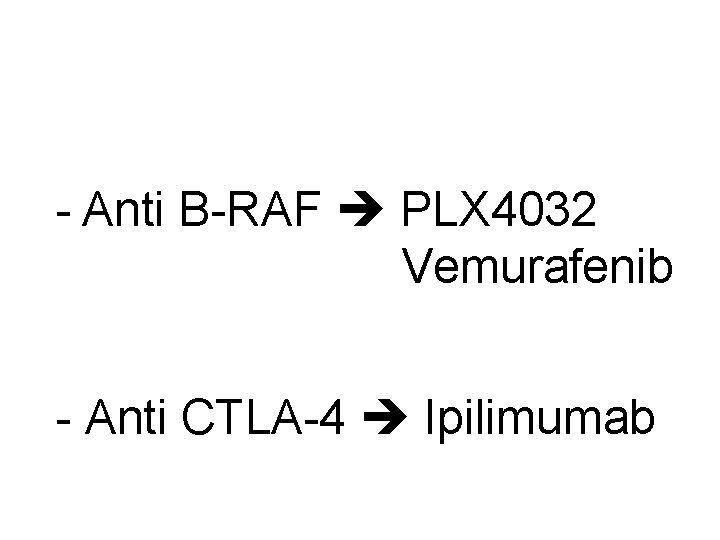
- Anti B-RAF PLX 4032 Vemurafenib - Anti CTLA-4 Ipilimumab

MOLECULAR PATHOLOGY IN MELANOMA • A change in the role of pathologist • Participation in multidisciplinary teams: – Her 2 neu and breast cancer – K-Ras and colon cancer – EGFR and pulmonary carcinoma – EML 4 -ALK and pulmonary carcinoma – Braf and metastatic melanoma

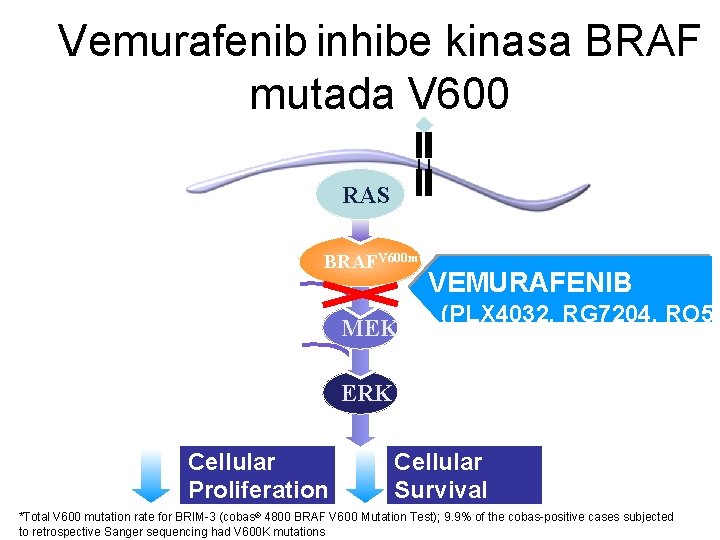
Vemurafenib inhibe kinasa BRAF mutada V 600 RTK RAS 50%* of melanomas BRAF RAFV 600 mut VEMURAFENIB ATP MEK (PLX 4032, RG 7204, RO 51 ERK Cellular Proliferation Cellular Survival *Total V 600 mutation rate for BRIM-3 (cobas® 4800 BRAF V 600 Mutation Test); 9. 9% of the cobas-positive cases subjected to retrospective Sanger sequencing had V 600 K mutations

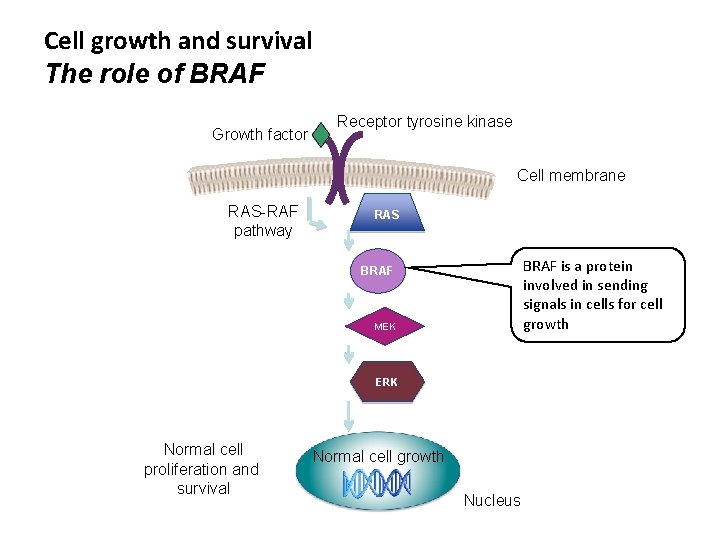
Cell growth and survival The role of BRAF Growth factor Receptor tyrosine kinase Cell membrane RAS-RAF pathway RAS BRAF is a protein involved in sending signals in cells for cell growth BRAF MEK ERK Normal cell proliferation and survival Normal cell growth Nucleus

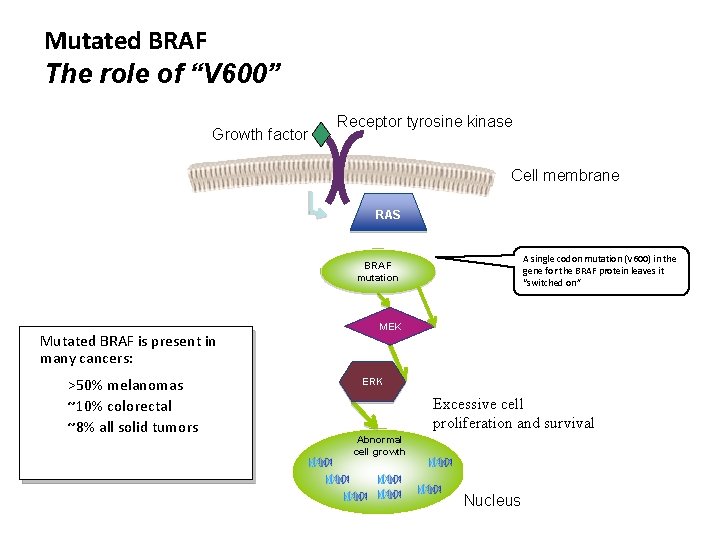
Mutated BRAF The role of “V 600” Growth factor Receptor tyrosine kinase Cell membrane RAS A single codon mutation (V 600) in the gene for the BRAF protein leaves it “switched on” BRAF mutation Mutated BRAF is present in many cancers: >50% melanomas ~10% colorectal ~8% all solid tumors MEK ERK Excessive cell proliferation and survival Abnormal cell growth Nucleus


CONCLUSIONS § The conventional H&E criteria are still the most useful in melanoma diagnostic and prognostic factors (Brelow I, ulceration, Growth phase). Brelow’s index is the most important prognostic factor §Some tools (tissue arrays, c. DNA arrays, RT-PCR) may be useful in order to discover new molecules that can help us to predict melanoma aggressiveness §Melanomas with Bcl 6 expression or loss p 16 have much more metastatic capability and kill the patient faster §Epithelium-mesenchymal transition molecular changes are directly involved in melanoma progression

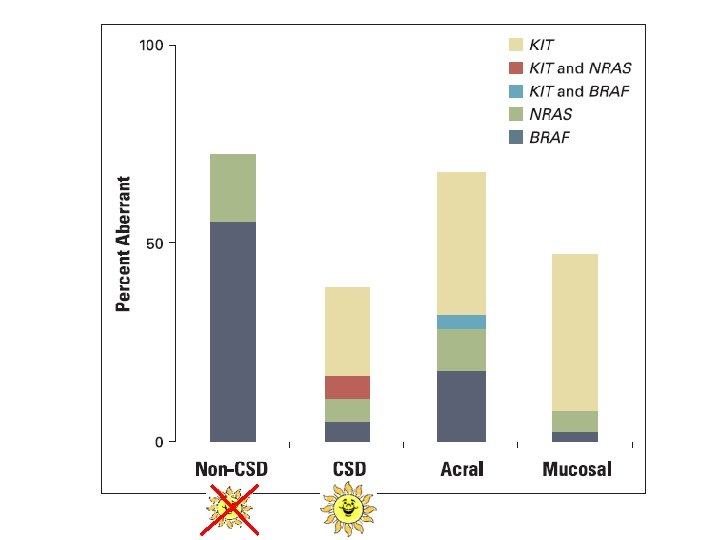
FUTURE § B-Rafomas: Those melanomas of Non chronic sun damage skin (trunk) susceptible to be treated with anti-BRAF drugs § C-Kitomas: Lentiginous Melanomas especially, lentigo maligna and acral lentiginous melanomas with c-Kit mutations § Gnaqomas: Ocular Melanomas (Gnaq-11 mutations)
