Molecular Pathological Classification Of Colorectal Cancer CRC Dr











- Slides: 39

Molecular Pathological Classification Of Colorectal Cancer (CRC) Dr. Calypso Barbatis MD, FRCPath, Ph. D HBD Histobiodiagnosis ATHENS HSGO / 19 -20 May 2018 ATHENS


Frequent malignancy 3 rd cause of cancer-related mortality worldwide Sporadic, Hereditary, Familial Related to IBD Precursor Lesions Almost perfect stepwise model of carcinogenesis Conventional adenoma Serrated lesions CRC


Specific features of CRC Heterogeneous Disease in morphology site gender age response to treatment Source: Pol J Path. 2014; 65(4): 257 -266. Bosman FT, Yan P.

Does CRC fit to the P 4 medicine Predictive Personalised Preventive Participatory Source: Hood L, Friend SH. Nat Rev Clin Oncol 2011; 8: 184 -187

Classical evidence-based Clinicopathological parameters Still determine treatment But The road to molecular classification has opened and awaits full implementation when clinical translation and precision medicine are approved

The classification of CRC is based on clinical morphological molecular features and available markers prognostic preventive

Current non-molecular classification of CRC Site Subsite (the most involved) Rectal: Inferior margin <16 cm from the anal verge or if any part of the tumor is located within the supply of the superior rectal artery

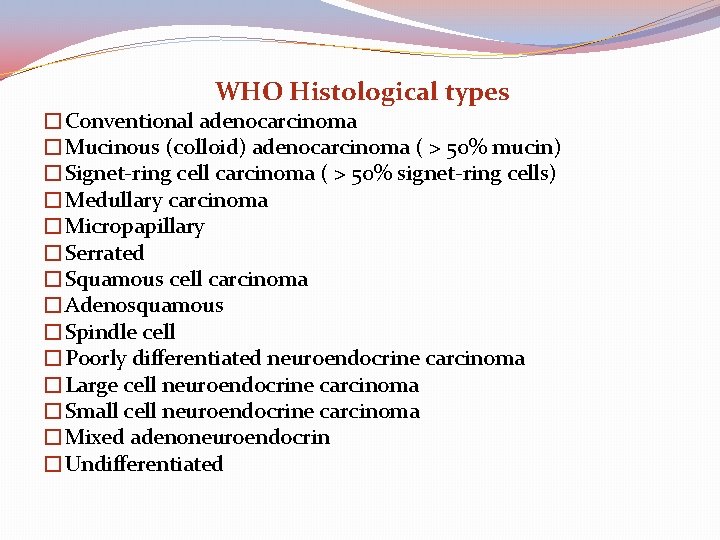
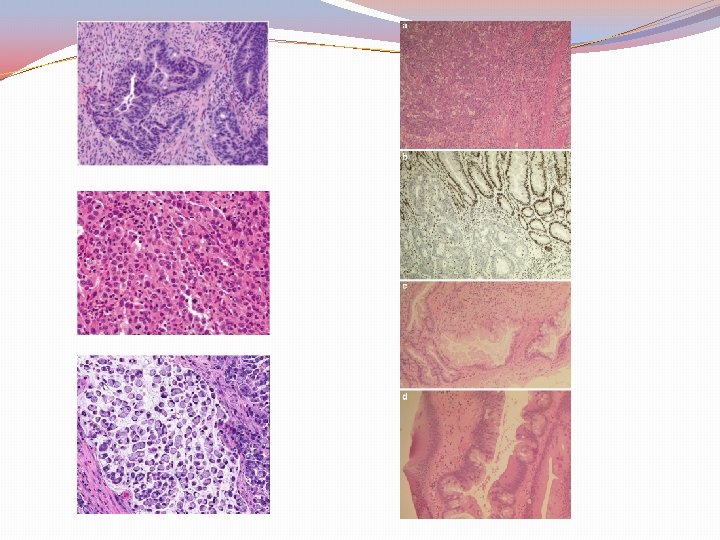
WHO Histological types �Conventional adenocarcinoma �Mucinous (colloid) adenocarcinoma ( > 50% mucin) �Signet-ring cell carcinoma ( > 50% signet-ring cells) �Medullary carcinoma �Micropapillary �Serrated �Squamous cell carcinoma �Adenosquamous �Spindle cell �Poorly differentiated neuroendocrine carcinoma �Large cell neuroendocrine carcinoma �Small cell neuroendocrine carcinoma �Mixed adenoneuroendocrin �Undifferentiated


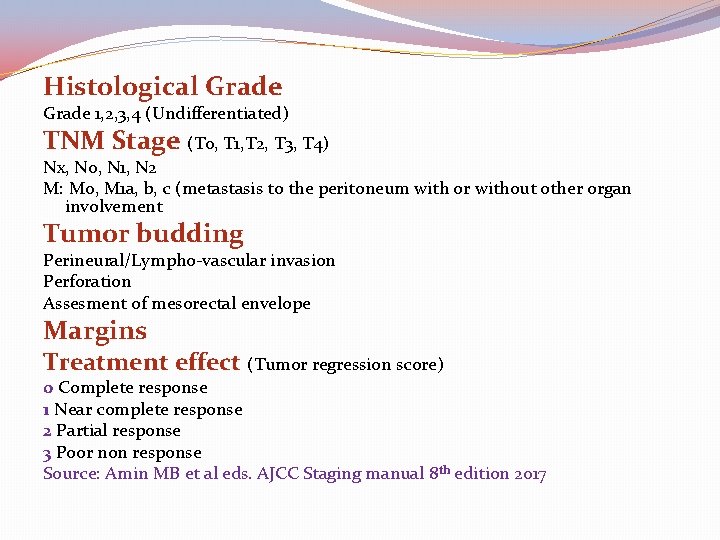
Histological Grade 1, 2, 3, 4 (Undifferentiated) TNM Stage (T 0, T 1, T 2, T 3, T 4) Nx, N 0, N 1, N 2 M: M 0, M 1 a, b, c (metastasis to the peritoneum with or without other organ involvement Tumor budding Perineural/Lympho-vascular invasion Perforation Assesment of mesorectal envelope Margins Treatment effect (Tumor regression score) 0 Complete response 1 Near complete response 2 Partial response 3 Poor non response Source: Amin MB et al eds. AJCC Staging manual 8 th edition 2017

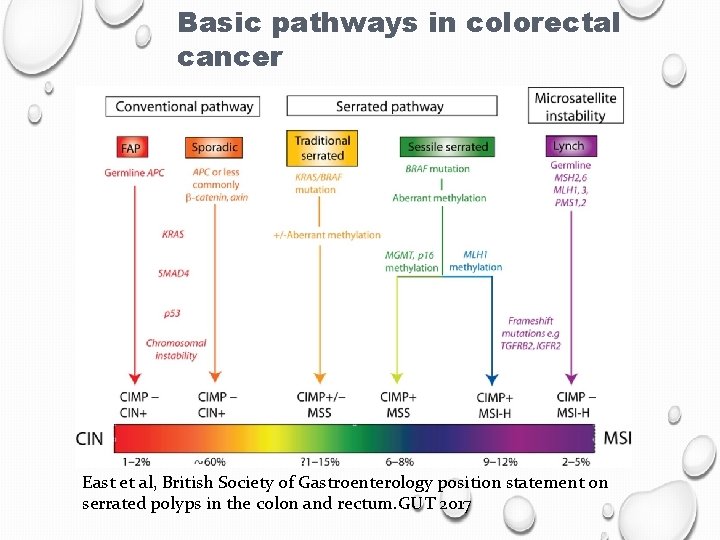
Basic pathways in colorectal cancer East et al, British Society of Gastroenterology position statement on serrated polyps in the colon and rectum. GUT 2017

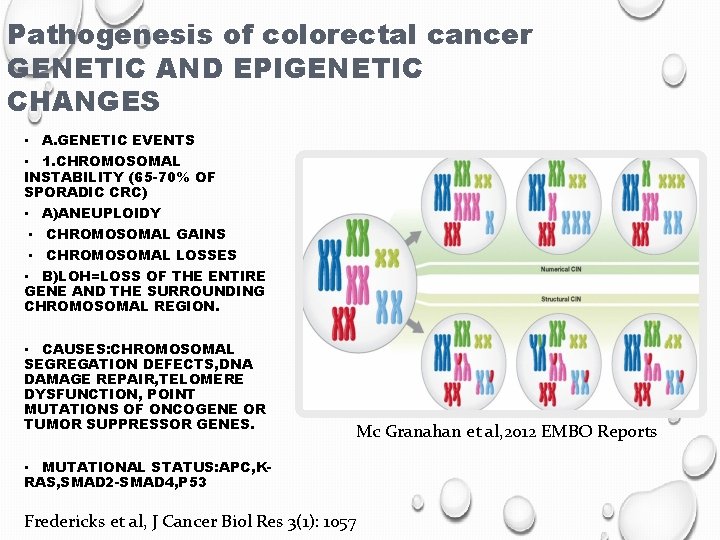
Pathogenesis of colorectal cancer GENETIC AND EPIGENETIC CHANGES • A. GENETIC EVENTS • 1. CHROMOSOMAL INSTABILITY (65 -70% OF SPORADIC CRC) • A)ANEUPLOIDY • CHROMOSOMAL GAINS • CHROMOSOMAL LOSSES • B)LOH=LOSS OF THE ENTIRE GENE AND THE SURROUNDING CHROMOSOMAL REGION. • CAUSES: CHROMOSOMAL SEGREGATION DEFECTS, DNA DAMAGE REPAIR, TELOMERE DYSFUNCTION, POINT MUTATIONS OF ONCOGENE OR TUMOR SUPPRESSOR GENES. Mc Granahan et al, 2012 EMBO Reports • MUTATIONAL STATUS: APC, KRAS, SMAD 2 -SMAD 4, P 53 Fredericks et al, J Cancer Biol Res 3(1): 1057

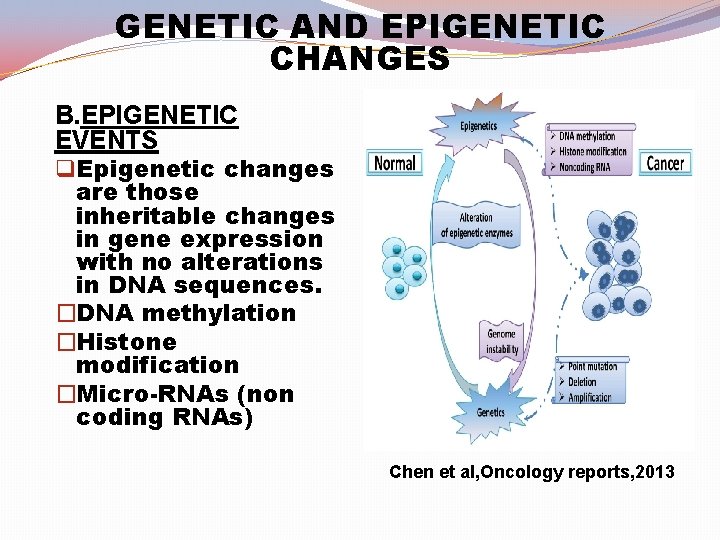
GENETIC AND EPIGENETIC CHANGES B. EPIGENETIC EVENTS q Epigenetic changes are those inheritable changes in gene expression with no alterations in DNA sequences. �DNA methylation �Histone modification �Micro-RNAs (non coding RNAs) Chen et al, Oncology reports, 2013

Molecular pathways of CRC carcinogenesis 1. Classical carcinogenetic model Chromosomal instability (CIN) Marked aneuploidy 1 st pathogenetic event APC gene mutations Allelic loss Somatic gene amplification Translocation Activation WNT signaling pathway

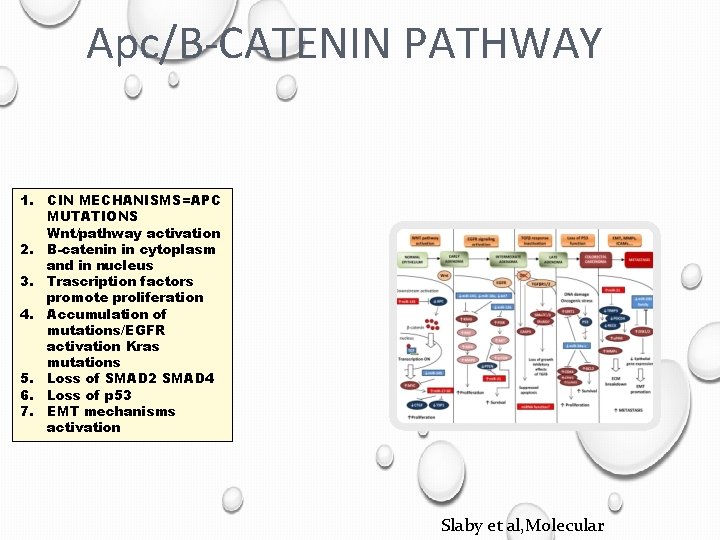
Apc/B-CATENIN PATHWAY 1. CIN MECHANISMS=APC MUTATIONS Wnt/pathway activation 2. B-catenin in cytoplasm and in nucleus 3. Trascription factors promote proliferation 4. Accumulation of mutations/EGFR activation Kras mutations 5. Loss of SMAD 2 SMAD 4 6. Loss of p 53 7. EMT mechanisms activation Slaby et al, Molecular

2 nd Pathway (MMR gene system or MSI) Inactivation of MMR genes Inactivation of suppressor genes Lynch syndrome (Diploid tumors with MSI)

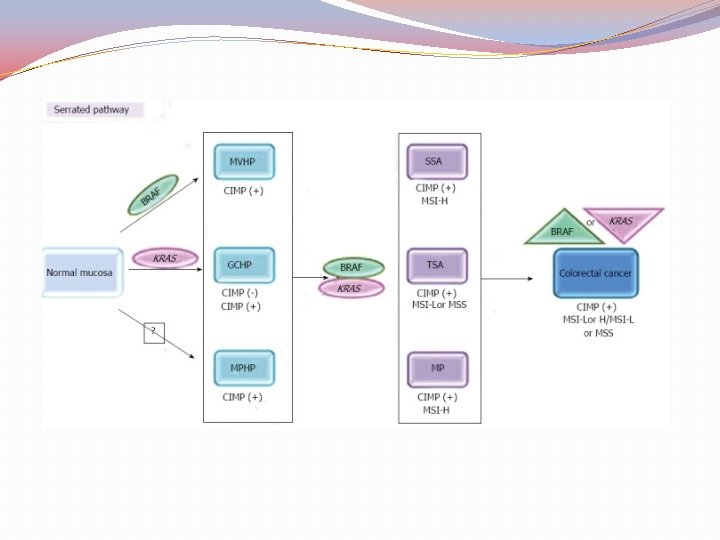
The Serrated pathway Hypermethylation of specific DNA regions near the “promoter genes” the Cp. G islands Diploid tumors Silence of Tumor Suppressor Genes


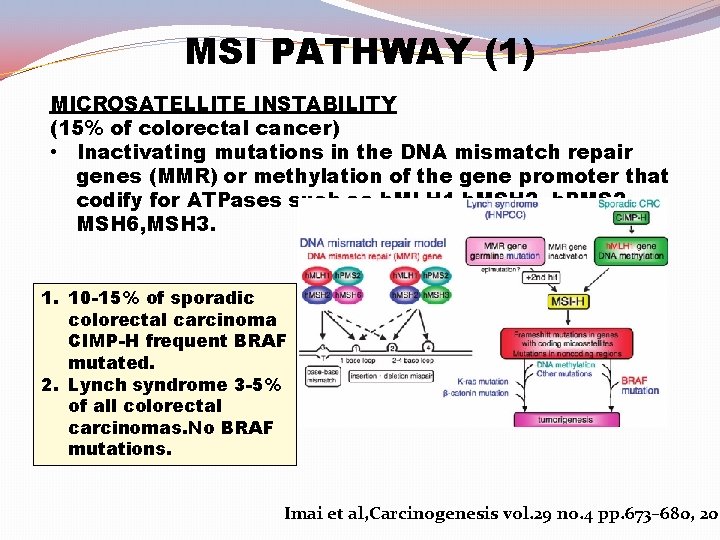
MSI PATHWAY (1) MICROSATELLITE INSTABILITY (15% of colorectal cancer) • Inactivating mutations in the DNA mismatch repair genes (MMR) or methylation of the gene promoter that codify for ATPases such as h. MLH 1, h. MSH 2 h. PMS 2, MSH 6, MSH 3. 1. 10 -15% of sporadic colorectal carcinoma CIMP-H frequent BRAF mutated. 2. Lynch syndrome 3 -5% of all colorectal carcinomas. No BRAF mutations. Imai et al, Carcinogenesis vol. 29 no. 4 pp. 673– 680, 200

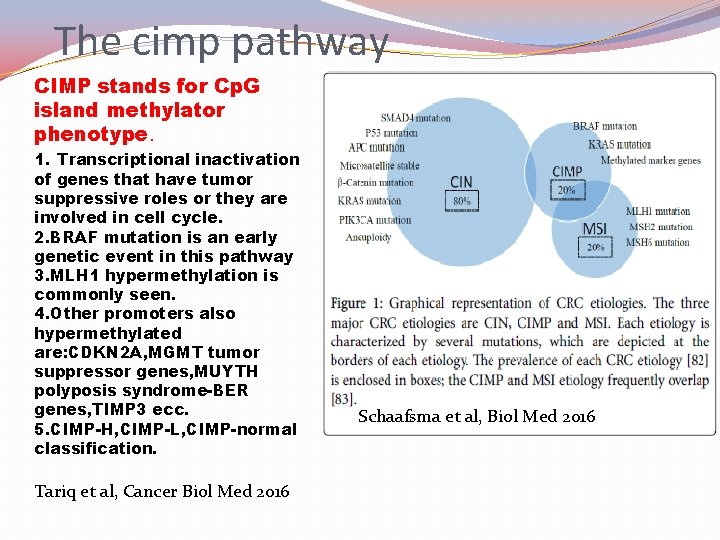
The cimp pathway CIMP stands for Cp. G island methylator phenotype. 1. Transcriptional inactivation of genes that have tumor suppressive roles or they are involved in cell cycle. 2. BRAF mutation is an early genetic event in this pathway 3. MLH 1 hypermethylation is commonly seen. 4. Other promoters also hypermethylated are: CDKN 2 A, MGMT tumor suppressor genes, MUYTH polyposis syndrome-BER genes, TIMP 3 ecc. 5. CIMP-H, CIMP-L, CIMP-normal classification. Tariq et al, Cancer Biol Med 2016 Schaafsma et al, Biol Med 2016

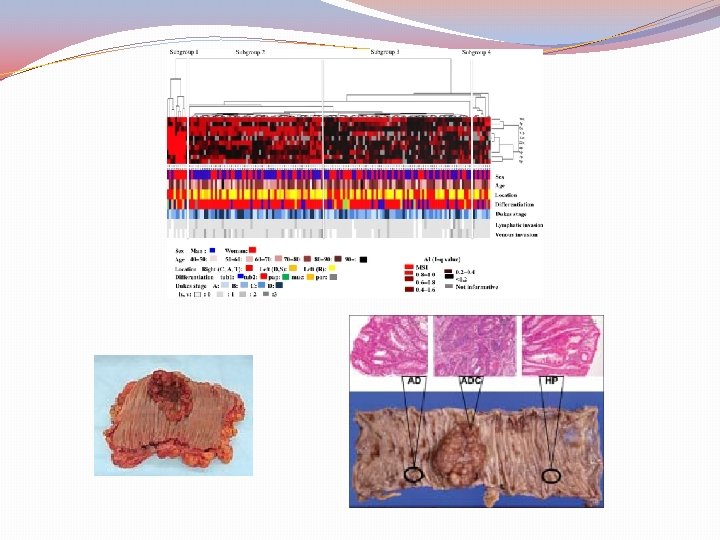
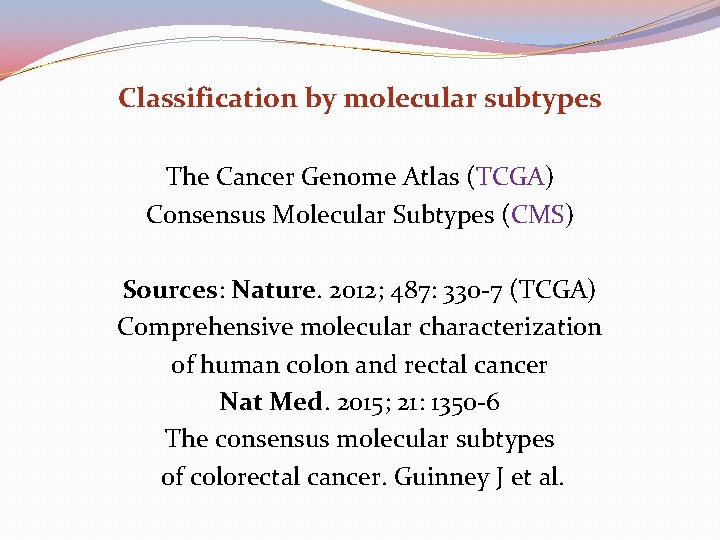
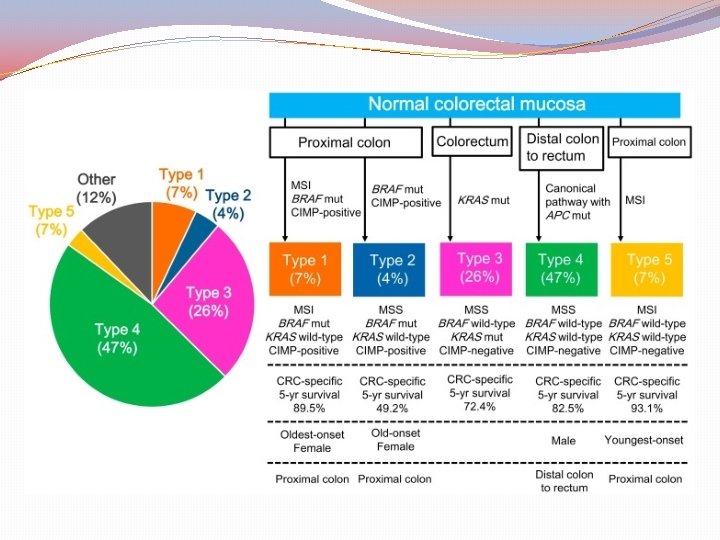
Classification by molecular subtypes The Cancer Genome Atlas (TCGA) Consensus Molecular Subtypes (CMS) Sources: Nature. 2012; 487: 330 -7 (TCGA) Comprehensive molecular characterization of human colon and rectal cancer Nat Med. 2015; 21: 1350 -6 The consensus molecular subtypes of colorectal cancer. Guinney J et al.

Purpose of classification “To correlate cancer cell phenotype with clinical behavior and provide guidance to rational treatment with specific targeted therapies” (CRC subtyping Concortium) This was attempted by the knowledge of gene expression data based on “epigenomic, transcriptomic, microenviromental, genetic and clinical characteristics of tumors’’



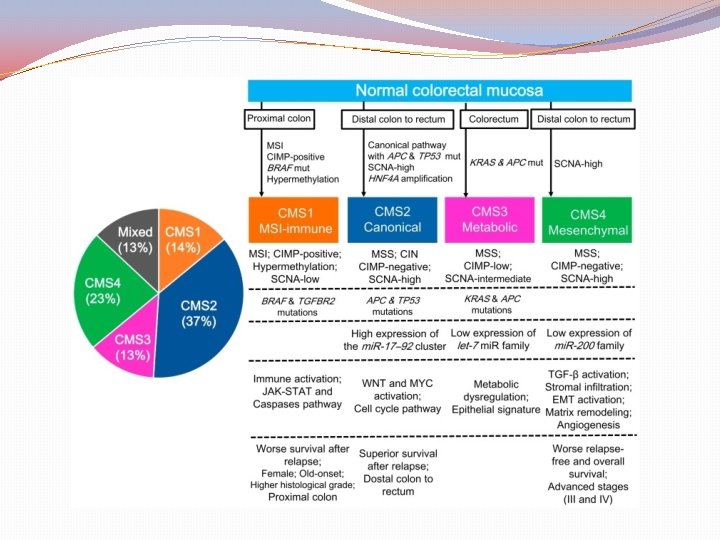
Consensus Molecular Subtypes of colorectal cancer (CMS) Assessment methods Mutations Chromosomal number alterations Methylation status Posttranslational Gene regulation Clinical analysis (site, gender, grade, stage) DFS (Disease Free Survival) RFS (Relapse Free Survival) SAR (Survival After Relapse) IMPORTANT!! “No subtype was solely defined by a genetic aberration” wild-type KRAS may be found in all subtypes

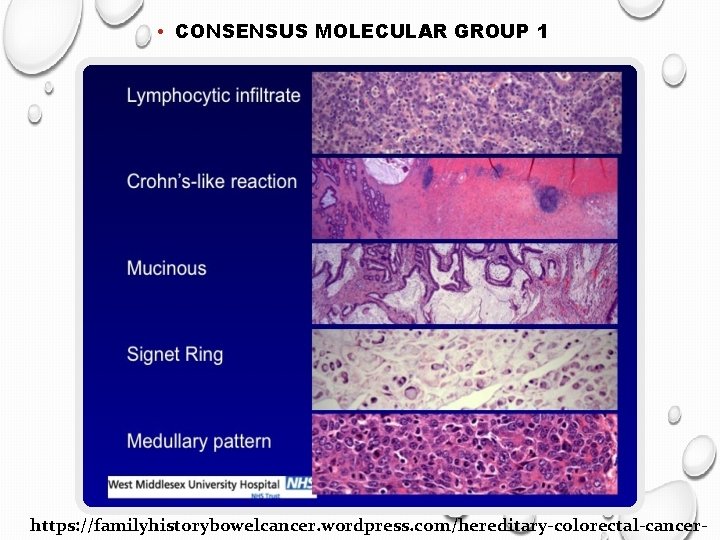
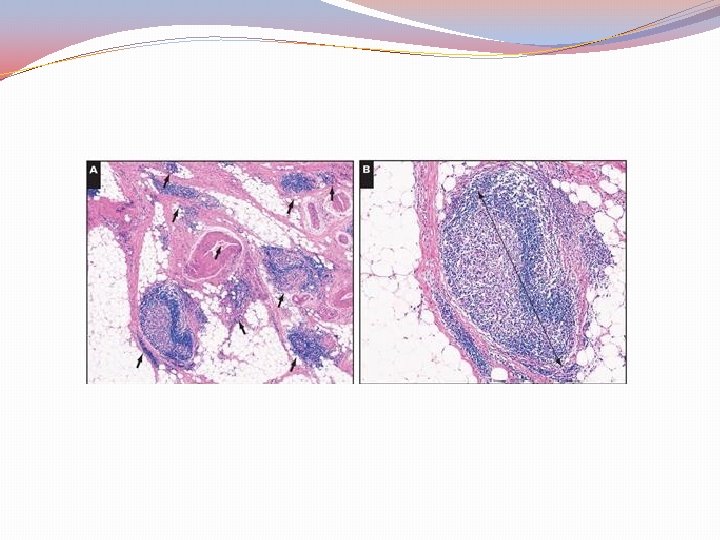
Clinical implications of CMS 1 (MSI) Better prognosis before dissemination Worst prognosis after relapse Stages 1, 2 perhaps no adjuvant chemotherapy Stage 3 no response to 5 -FU Importance of the immunogenic microenvironment Treatment? Check point inhibitors

• CONSENSUS MOLECULAR GROUP 1 https: //familyhistorybowelcancer. wordpress. com/hereditary-colorectal-cancer-


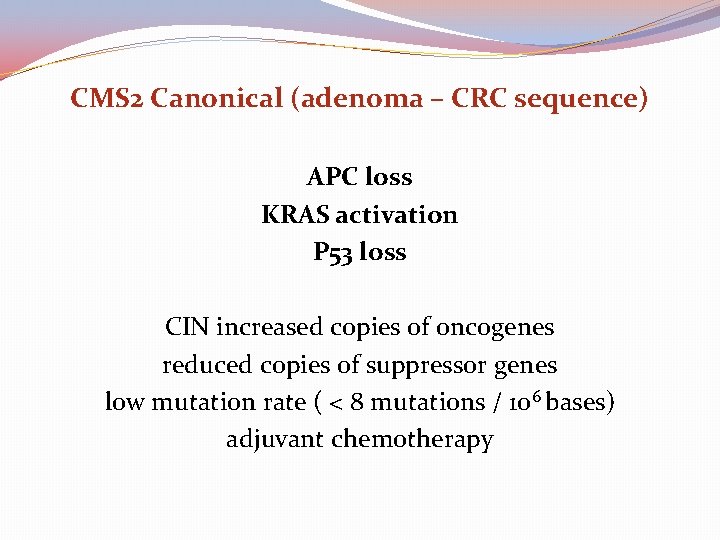
CMS 2 Canonical (adenoma – CRC sequence) APC loss KRAS activation P 53 loss CIN increased copies of oncogenes reduced copies of suppressor genes low mutation rate ( < 8 mutations / 106 bases) adjuvant chemotherapy

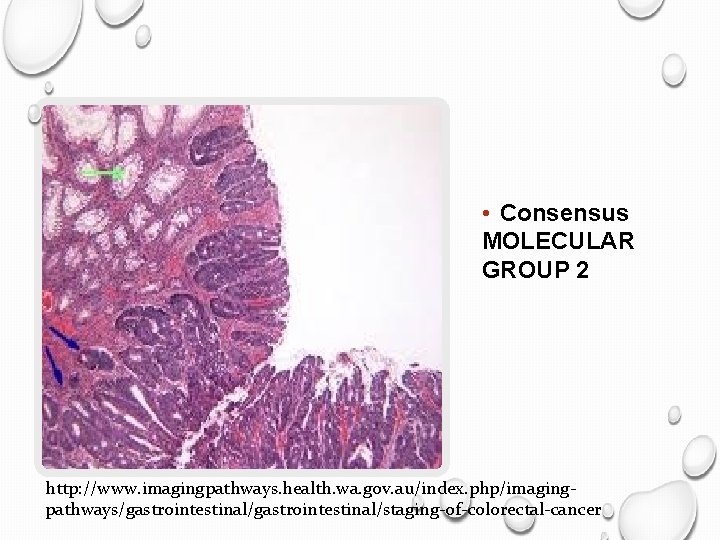
• Consensus MOLECULAR GROUP 2 http: //www. imagingpathways. health. wa. gov. au/index. php/imagingpathways/gastrointestinal/staging-of-colorectal-cancer

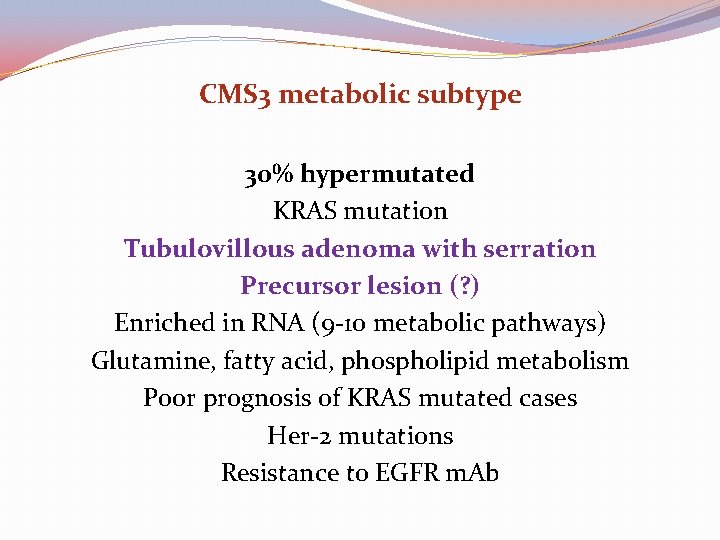
CMS 3 metabolic subtype 30% hypermutated KRAS mutation Tubulovillous adenoma with serration Precursor lesion (? ) Enriched in RNA (9 -10 metabolic pathways) Glutamine, fatty acid, phospholipid metabolism Poor prognosis of KRAS mutated cases Her-2 mutations Resistance to EGFR m. Ab

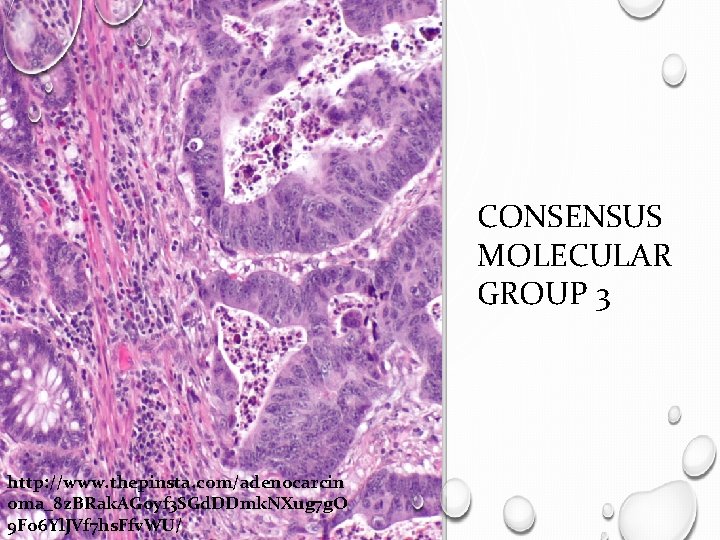
CONSENSUS MOLECULAR GROUP 3 http: //www. thepinsta. com/adenocarcin oma_8 z. BRak. AGoyf 3 SGd. DDmk. NXug 7 g. O 9 Fo 6 Yl. JVf 7 hs. Ffv. WU/

CMS 4 (mesenchymal) Advanced stage in diagnosis Poor survival (62% in 5 years) Resistant to anti-EGFR Regardless of KRAS mutation status Difficult to treat ? use of microenvironment features ? anti avβ 6 integrin Anti-CAF Anti-macrophage Source: Thanki K et al. IBBJ 2017; 3(3) 105 -111

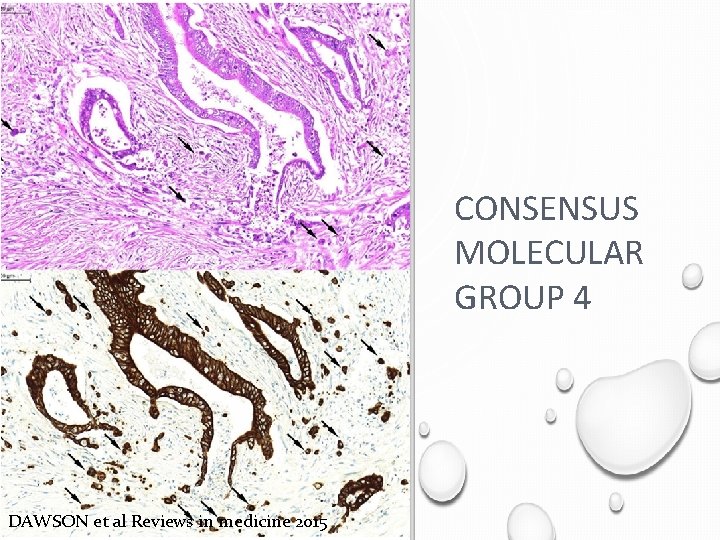
CONSENSUS MOLECULAR GROUP 4 DAWSON et al Reviews in medicine 2015

Molecular features of CRC Molecular Heterogeneity Genomic, epigenomic define Molecular subtypes Implementation of Personalized therapies Improvement of management

Acknowledgement My gratitude to Dr. Bouklas for providing some of the tables and additional literature

Thank You For Your Attention