Molecular Modeling in the Undergraduate Curriculum Molecular Modeling















- Slides: 44

Molecular Modeling in the Undergraduate Curriculum

Molecular Modeling in the Undergraduate Curriculum Rebecca R. Conry l Shari U. Dunham l Stephen U. Dunham l Margaret H. Hennessy l D. Whitney King l Julie T. Millard l Bradford P. Mundy l Dasan M. Thamattoor l Thomas W. Shattuck l -Inorganic -Biochemistry -Physical -Analytical -Biochemistry -Organic -Physical

Acknowledgements l National Science Foundation 1993, 1996 l Howard Hughes Medical Institute 1996, 2000 l Paul J. Schupf Scientific Computing Center 1993, 1996, 1998 l New England Consortium for Undergraduate Science Education 1998

Introduction-Molecular Modeling Information Technology l Stereo. Visualization - Molecular Perspective l Build Insight to solve challenges l Unified array of techniques l Link lecture and laboratory l Where, When, and How l

When, Where, How? Research focus l Teaching and research boundaries blur l Time is short: expertise and skills l Research quality tools early l Repeat Use Often l Productive student research l

When, Where, How Classroom l Laboratory l Homework l Projects l Tests l Thesis l

Information Rich Curriculum Massive amount of information l Database technology l Central to mission of the course l Students are active participants l » Gathering and Assessing Quality l Simulation and Prediction

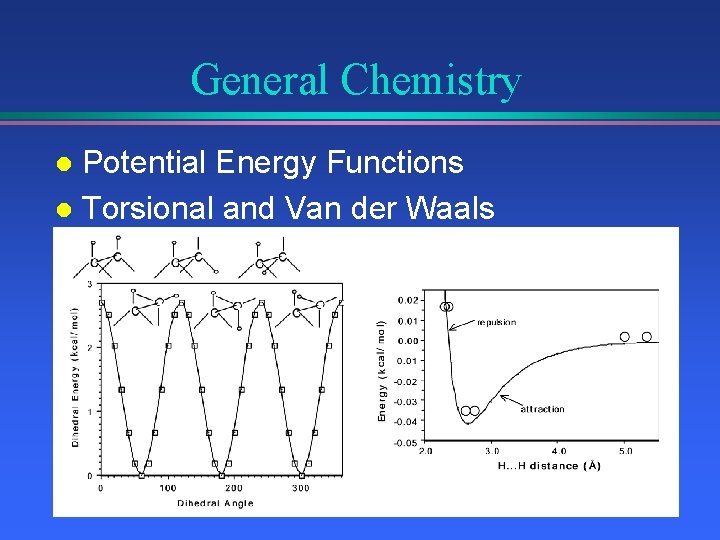
General Chemistry Potential Energy Functions l Torsional and Van der Waals l

General Chemistry l Molecular Mechanics » Hyroxyl Group » H 2 O 2, Methanol, Amino Acids » Catalase, Alcohol Dehydrogenase » Insight/CHARMm l Molecular Orbital Theory » O 2 MO’s (Sontum, Walstrum, Jewett) » Electrostatic Distributions (Shusterman) » Spartan

Molecular Structure Calculations 800 careful calculations on small inorganics and organics l Density functional theory gets MO ordering right for diatomics l NBO analysis for best Lewis Structure l » localized electron pair model » hybridization l Searchable Web database

Molecular Structure Calculations

Organic Radical Cations, Neutral Radicals, Cations, and Anions ethyl*

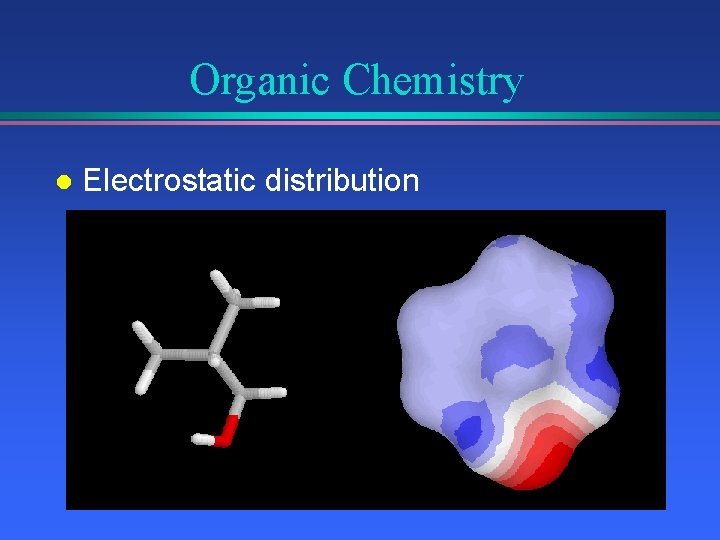
Organic Chemistry l Electrostatic distribution

Organic Chemistry l Molecular Orbital Theory- Spartan » Dimethylhexanes- ring conformations » Stabilities of butyl cations » Electrostatic distribution in allyl ions l Independent Projects » Synthesis and computation component » reaction intermediates » isomer energies » Dilantin, Strawberry Aldehyde, Limonene

Organic Chemistry l Bridgehead alkenes and cations

Physical Chemistry l Molecular Mechanics » Insight/CHARMm, MM 2 Molecular dynamics l Free Energy Perturbation Theory l » solvation » binding eguilibrium l Molecular Orbital Theory » Spartan, MOPAC, Gaussian

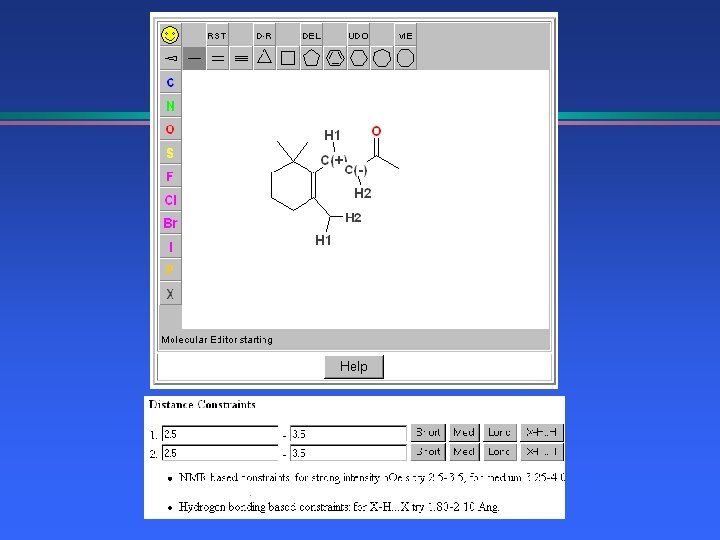
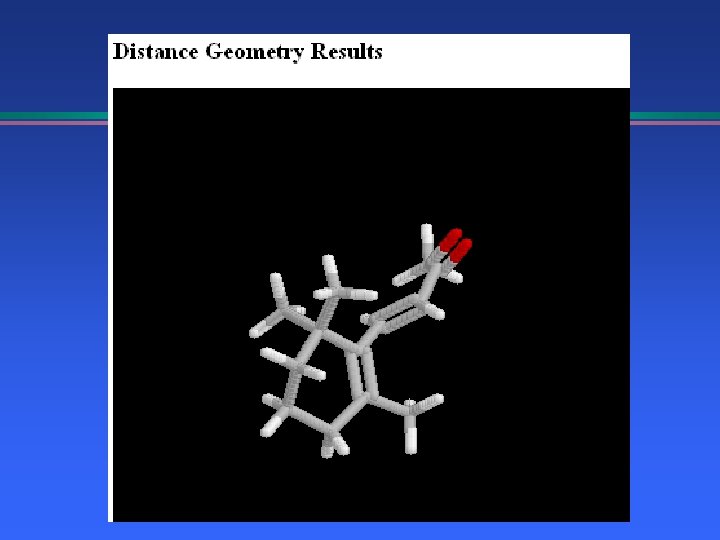
NMR and Distance Geometry Beta-ionone side chain geometry l T 2 relaxation; COSY, NOESY 2 D-NMR l Molecular Mechanics l Molecular Dynamics l » Correlate motion with relaxation times » Explore conformation space l Distance Geometry based on n. Oe’s

Distance Geometry NMR Constraints, n. Oe distances l Hydrogen bonding constraints l Generate 3 D structure l 2 D->3 D conversion l Follow with Molecular Mechanics l » EMBED: G. Crippen, I. Kuntz, T. Nordland, T. Havel, UCSF » Java. Molecular. Editor, Peter Ertl at Novartis



Gramicidin-S Open ended student project l secondary and tertiary structure l COSY, NOESY 2 D-NMR l Molecular Mechanics, constrained by n. Oe and hydrogen bonding constraints l Molecular Dynamics l

Computer Aided Molecular Design A Strategy for Meeting the Challenges We Face

An Organized Guide Build Chemical Insight l Discover new molecules l Predict their properties l

Principles Structure-Function Relationships l Binding l » Understand control binding ->disease l Molecular Recognition » How do enzymes recognize and bind the proper substrates l Guest-Host Chemistry » Molecular Recognition in Cyclodextrins

Hosts: cyclodextrin

CAMD Determine Structure of Guest or Host l Build a model of binding site l Search databases for new guests (or hosts) l Dock new guests and binding sites l Predict binding constants or activity l Synthesize guests or hosts l

Binding Site Model Using experimental binding constants l Build interaction model of binding site l Use 3 D database searching to find other tight-binding guests l

Structure Searches 2 D Substructure searches l 3 D Conformationally flexible searches l » cfs

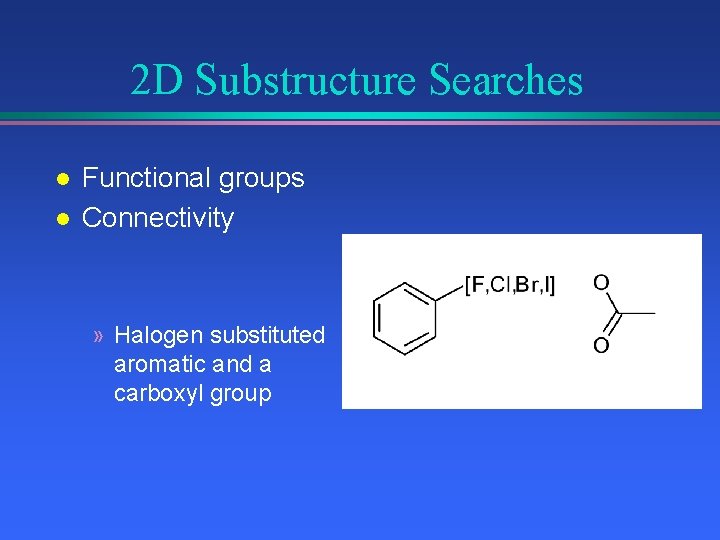
2 D Substructure Searches l l Functional groups Connectivity » Halogen substituted aromatic and a carboxyl group

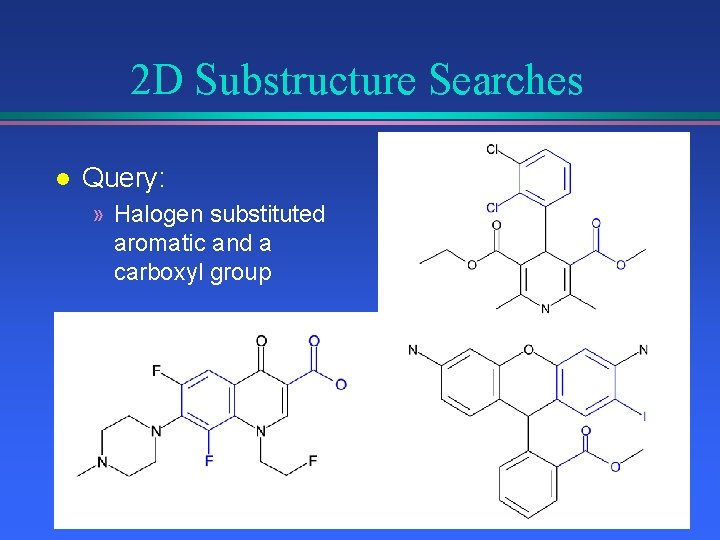
2 D Substructure Searches l Query: » Halogen substituted aromatic and a carboxyl group

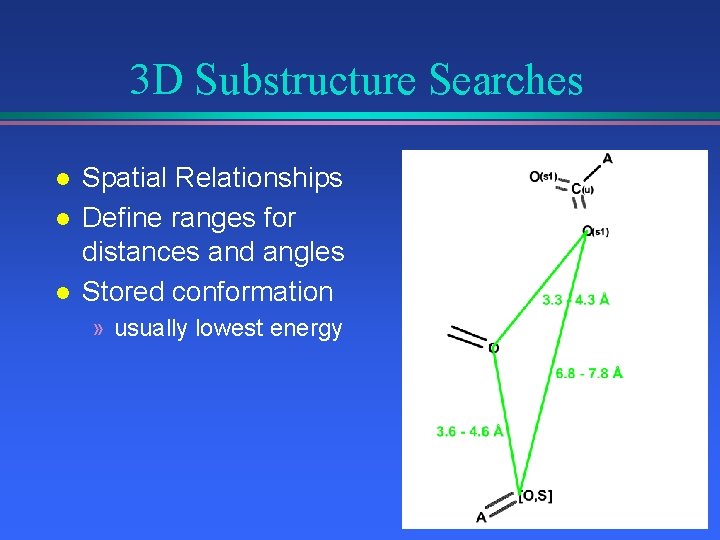
3 D Substructure Searches l l l Spatial Relationships Define ranges for distances and angles Stored conformation » usually lowest energy

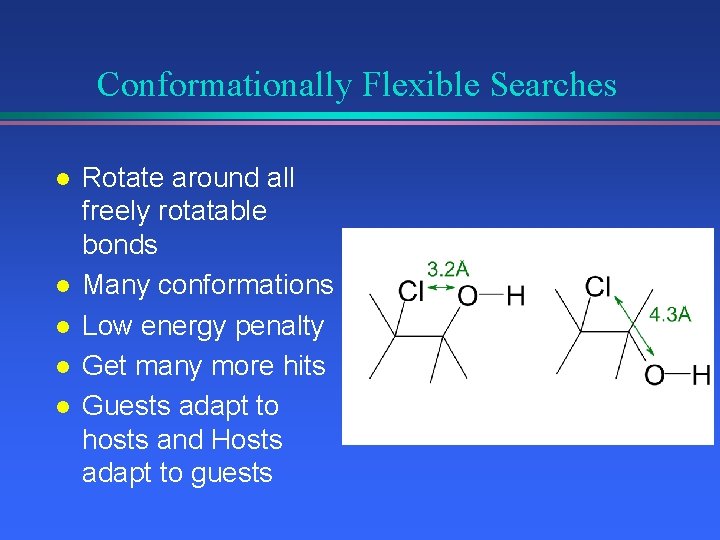
Conformationally Flexible Searches l l l Rotate around all freely rotatable bonds Many conformations Low energy penalty Get many more hits Guests adapt to hosts and Hosts adapt to guests

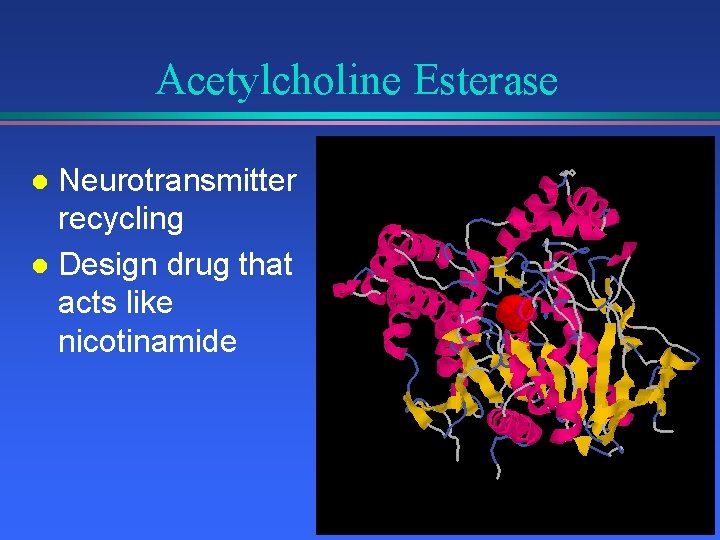
Acetylcholine Esterase Neurotransmitter recycling l Design drug that acts like nicotinamide l

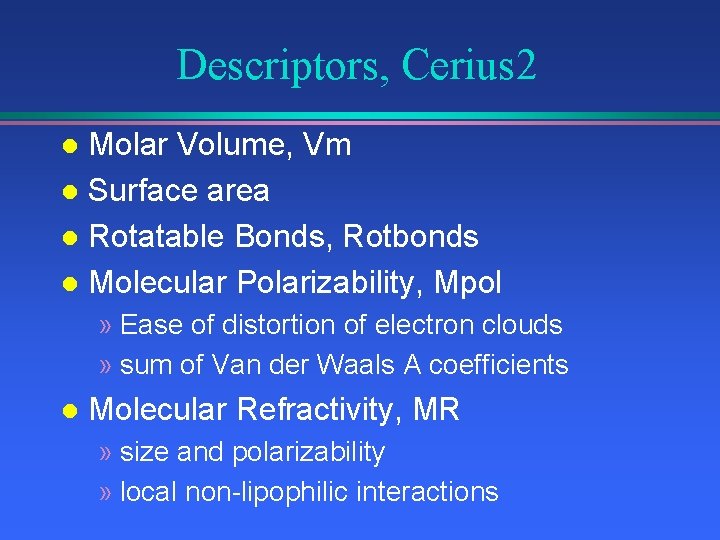
Descriptors, Cerius 2 Molar Volume, Vm l Surface area l Rotatable Bonds, Rotbonds l Molecular Polarizability, Mpol l » Ease of distortion of electron clouds » sum of Van der Waals A coefficients l Molecular Refractivity, MR » size and polarizability » local non-lipophilic interactions

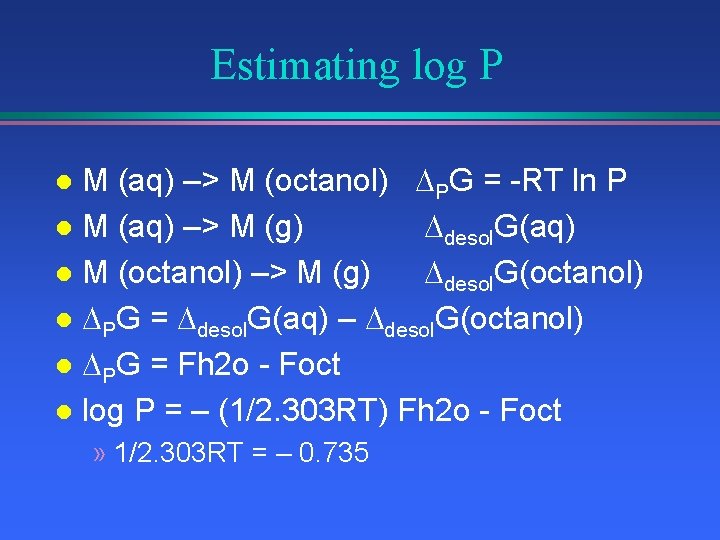
Estimating log P M (aq) –> M (octanol) PG = -RT ln P l M (aq) –> M (g) desol. G(aq) l M (octanol) –> M (g) desol. G(octanol) l PG = desol. G(aq) – desol. G(octanol) l PG = Fh 2 o - Foct l log P = – (1/2. 303 RT) Fh 2 o - Foct l » 1/2. 303 RT = – 0. 735

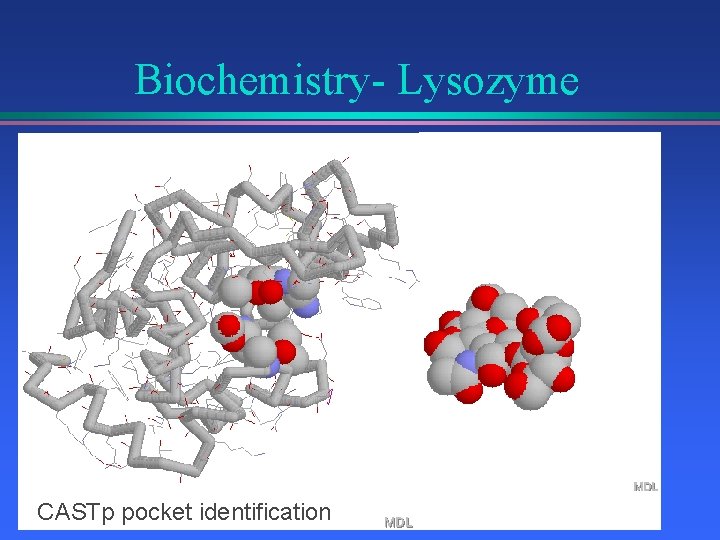
Biochemistry- Lysozyme CASTp pocket identification

Biochemistry Conformational Energetics of Oligosaccharides l Stereospecificity of Lactate Dehydrogenase Isozymes l Bioinformatics l Homology Modeling l

ENZYME Monoamine oxidase A Search in ENZYME for: monoamine oxidase a 1. 4. 3. 4 Amine oxidase (flavin-containing). (AN: Monoamine oxidase. Tyraminase. Amine oxidase. Adrenalin oxidase. )

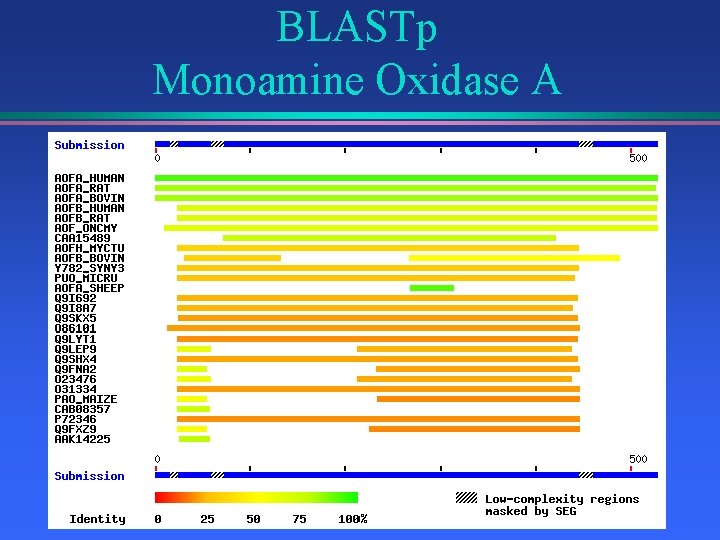
BLASTp Monoamine Oxidase A

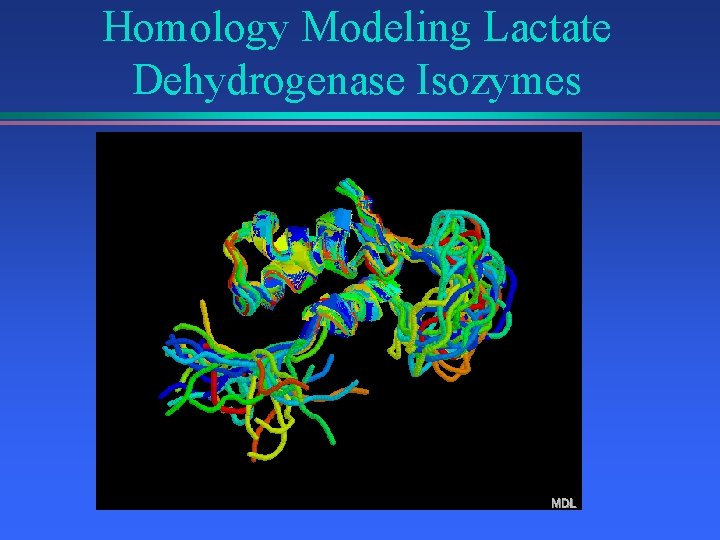
Homology Modeling Lactate Dehydrogenase Isozymes

Synthesis- Reaction Databases 600, 000 organic reactions l Synthetic routes l Chem. Inform - Reacts, ISIS/Host l Access by substructure and bond rearrangments l Most used database by students l

High Throughput Screening Test 10, 000 -100, 000’s of compounds l Robotics l » Individually tested » Pfizer: > 250, 000 compound library l Combinatorial Chemistry » Parallel testing » Cleverly prepared mixtures » Recover most active compounds

Proteomics LC/MS - Bioinformatics l Protein complement (30, 000 -60, 000) l » Expression proteomics Localization within cell l Protein interactions l » Interaction proteomics l Database searching » Sequence tag-Tag. Ident » MS/MS-Sequest

Making Room Have no choice l Student independent research expected l » job interviews » best grad schools l Student perceptions: teach more » enlivens classroom » relevant » build expertise l Wet/Lab Computational lab