Molecular Marker Characterization of plant genotypes Morphological markers












- Slides: 30

Molecular Marker Characterization of plant genotypes Morphological markers Physiological markers Biochemical markers Molecular markers etc.

Widely-used markers To distinguish varieties / genotypes by observation / measurement Characteristics: growth habit, fruit color, shape, etc. resp rate, PS content, hormone balance, etc. fruit size, plant height, sugar content, etc.

Molecular Marker Useful when other methods not available / possible Very similar morphology / anatomy Growth and development stages Environmental factors Analysis of banding patterns Statistics for evaluation of polymorphism

Molecular marker Study and management of genetic resources Identifying and distinguishing genotypes Marker assisted selection (MAS) Complementary tool for DUS studies of cultivars Distinctiveness / Uniformity / Stability

Molecular markers Protein-based marker: Isozyme / Allozyme multiple molecular forms of an enzyme similar / identical catalytic activity enzyme assay on PAGE DNA-based marker

Isozyme marker

Isozyme marker Enzyme Isozyme locus # allozymes Shikimate dehydrogenase Sdh-1 1 Phosphoglucose isomerase Pgi-1 2 Pgi-2 2 Aminonentidase Amp-1 3 Amp-2 4 Amp-3 3 Amp-4 4 Alcohol dehydrogenase Adh-1 2 Phosphogluconate dehydrogenase Pgd-1 2 Pgd-2 1 Glu oxaloacetate transaminase Got-1 1 Got-3 1

DNA-based markers Approach: Hybridization Polymerase Chain Reaction Types: RFLP Minisatellite RAPD SCAR SSR AFLP SNP etc

DNA-based markers Patterns of small DNA sequences Constant landmarks in the genome May or May not have biological function Linked to conserved or variable regions

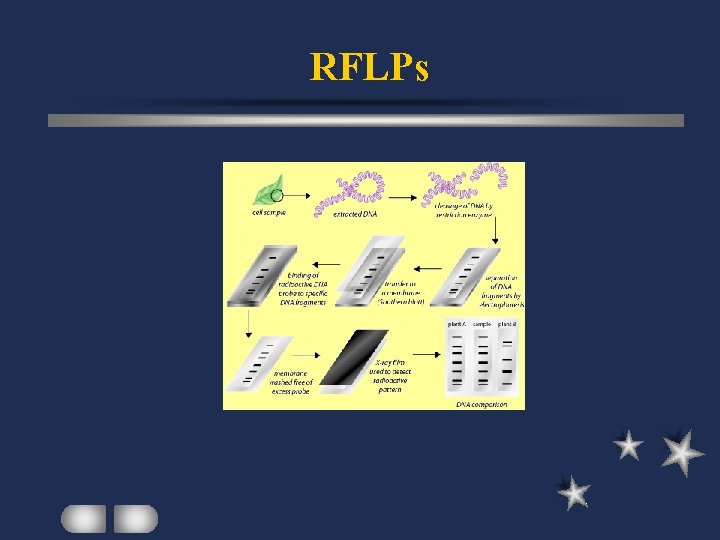
RFLPs Restriction Fragment Length Polymorphisms Digestion with restriction enzymes Size fractionation on agarose gel Southern hybridization (genomic or c. DNA probe) Analysis of hybridized restriction fragments

RFLPs Polymorphism: homologous pieces of different lengths mutation on restriction sites mutation between restriction sites

RFLPs Several bands per lane Highly polymorphic in a population at a locus – max 2 alleles in an individual Co-dominant marker Laborious / Time consuming Usually use isotope

RFLPs

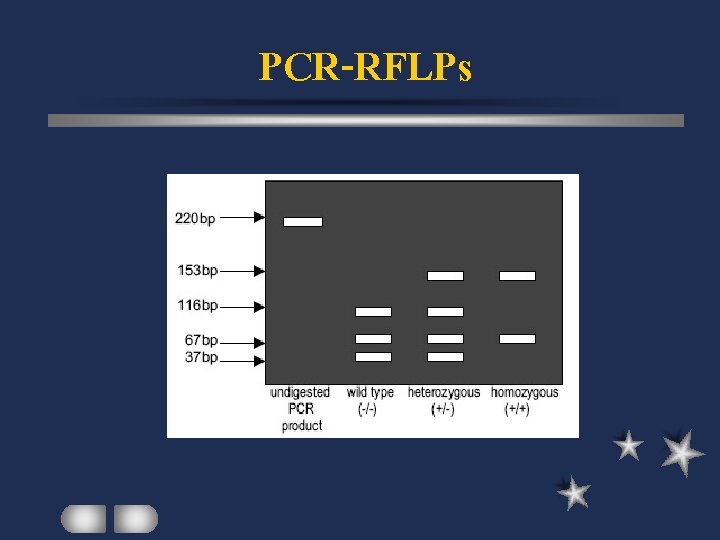
PCR-RFLPs

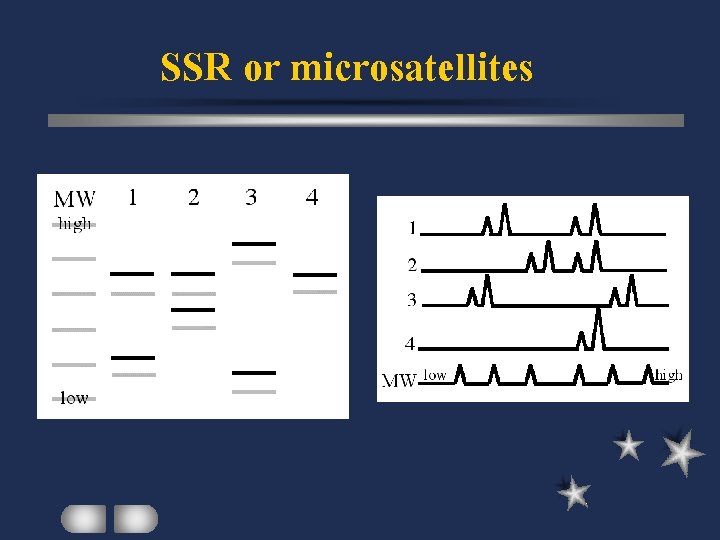
SSR or microsatellites Simple Sequence Repeat several bases per repeat tandem repeats flanked by unique sequences primer design based on flanking sequences polymorphism: number of repeating units

SSR or microsatellites Easy to detect via PCR High polymorphism Co-dominant marker Library screening or Database search require for sequence identification

SSR or microsatellites

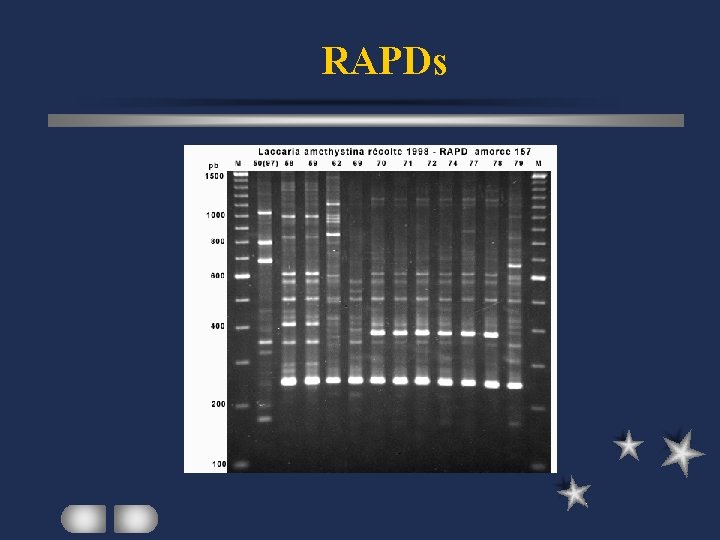
RAPDs Random Amplified Polymorphic DNAs PCR with 1 short primer (usu decamer) low annealing temperature primer annealing in inverted orientation at optimal distances amplified products analyzed on agarose gel

RAPDs Polymorphisms: base changes at annealing sites insertion/deletion within amplified fragments Results: presence or absence of the bands Cannot distinguish homozygote / heterozygote

RAPDs Simple, fast, relatively inexpensive assay Many loci to be identified in 1 rxn Can be automated Inconsistent results (short primer / low temp) Less informative for mapping with dominant nature different lengths not identifiable

RAPDs

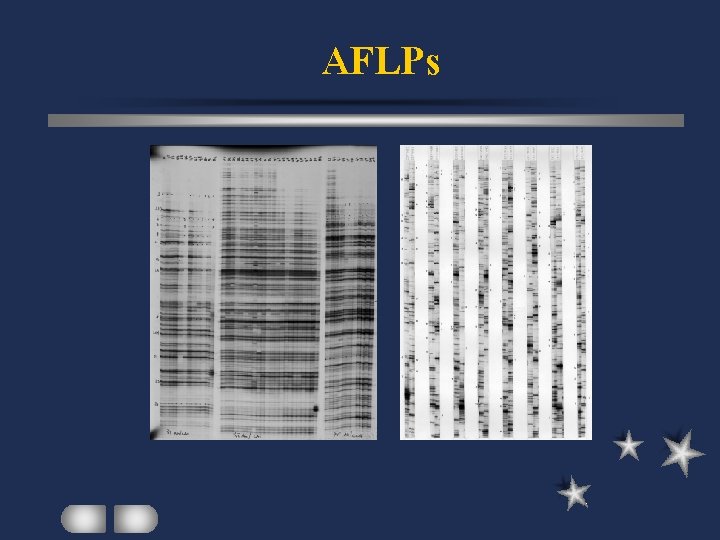
AFLPs Amplified Fragment Length Polymorphisms digestion with 2 enzymes (rare/frequent cutters) eg Eco. RI and Mse. I ligation of synthetic adapters to RFs pre-selective amplification primers corresponding to adapter sequences

AFLPs Amplified Fragment Length Polymorphisms selective amplification 1 -5 nt added to 3’ end of each primer 1 nt added to each primer 1/16 amplified banding patterns analyzed by PAGE

AFLPs Many loci to be identified in 1 rxn High efficiency in detecting polymorphic DNA More consistent pattern than RAPDs Dominant marker Technically challenging / labor intensive

AFLPs

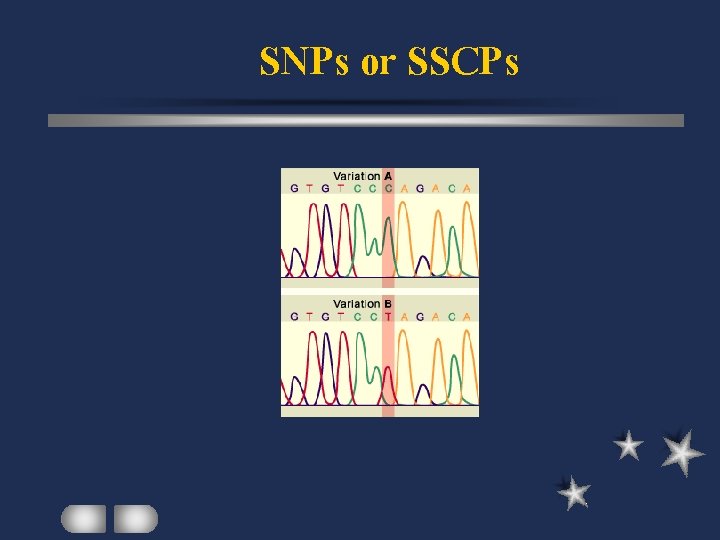
SNPs or SSCPs Single Nucleotide Polymorphisms Single-Stranded Conformation Polymorphisms SNP: major genetic source of phenotypic variability differentiate individuals within a species

SNPs or SSCPs Mobility of ss. DNA dependent of nt sequence looping or compaction Polymorphisms at a single locus base change by point mutation or small insertion / deletion

SNPs or SSCPs Specific primers to amplify target region Asymmetric PCR (1 primer in excess) Regular PCR (denaturing ds DNA) ss PCR products analyzed by electrophoresis Base change revealed by labeled nucleotides in automated sequencer

SNPs or SSCPs Many approaches for detection PCR-RFLP primer extension allele specific oligonucleotide ligation allele specific hybridization sequencing

SNPs or SSCPs