Molecular genetics Previous discussions focused on the individual





- Slides: 23

Molecular genetics • Previous discussions focused on the individual. • Focus has now shifted to genes • • • How are they encoded (3/29, 4/2) How are they replicated (4/2, 4/5) How are they expressed - transcription (4/7) How are they expressed - translation (4/9) Relationship between phenotype and genotype - pathways (4/12) • How are they regulated (4/14, 4/16, 4/19) • How we study them - individual genes (4/19, 4/21, 4/23) • Review 4/28 and Exam 4/30 • How we study them - global studies (Genomics-4/26, 5/3, 5/5) • Final Exam 5/10

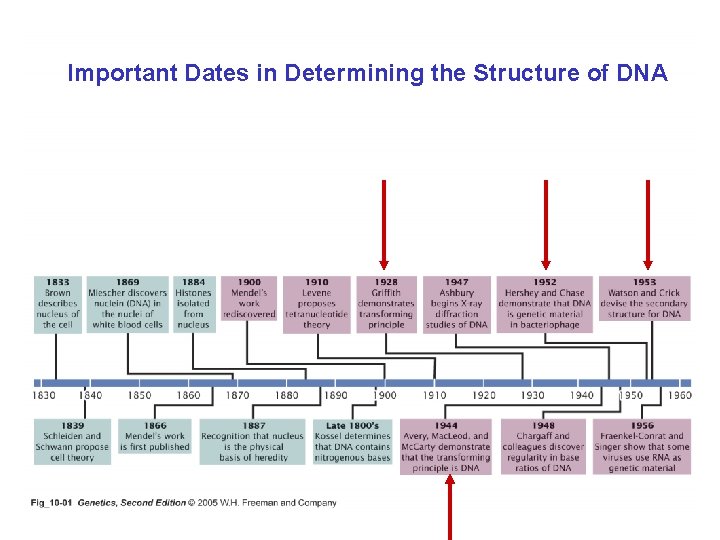
Important Dates in Determining the Structure of DNA

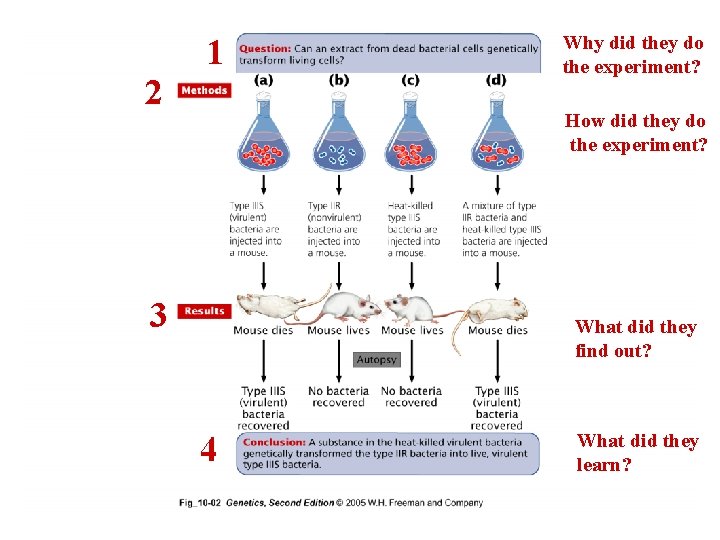
1 2 Why did they do the experiment? How did they do the experiment? 3 What did they find out? 4 What did they learn?

Model question • One interpretation of the results obtained by Griffith after performing experiment (d, previous slide) is stated as the “conclusion” in previous slide. State one other possible interpretation? What experiments and conclusion did they use to rule out this interpretation?

Review the findings that were used by Watson and Crick to propose the double helix model for the structure of DNA – Erwin Chargaff’s data – Rosiland Franklin’s x-ray crystallography data

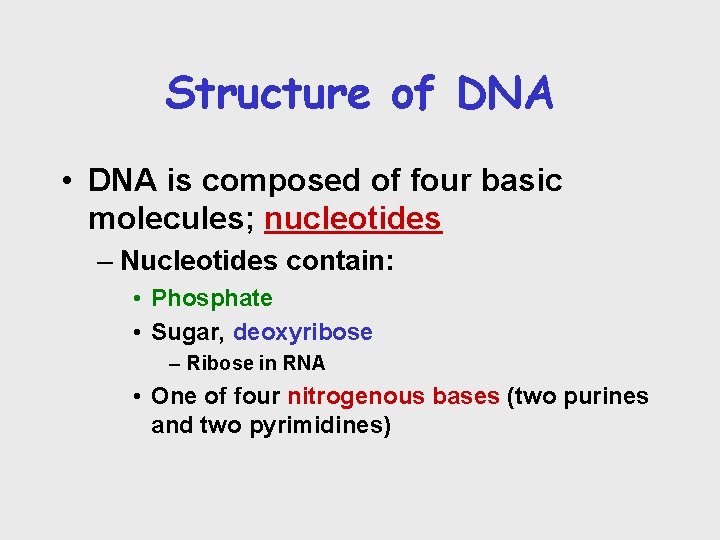
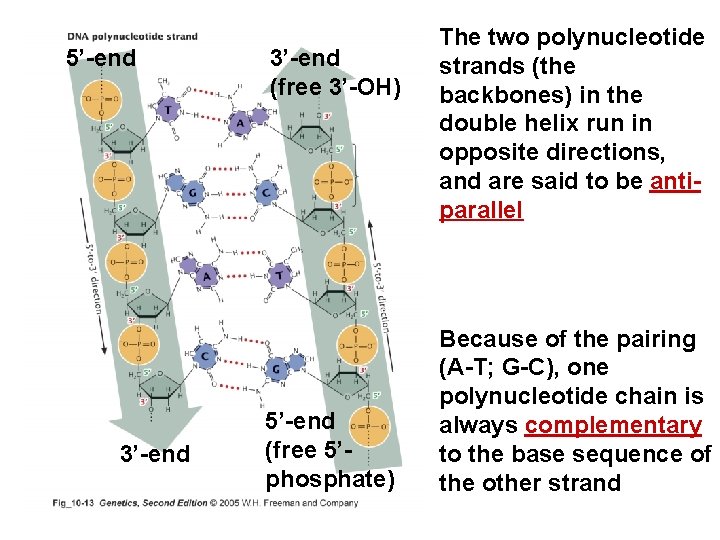
Structure of DNA • DNA is composed of four basic molecules; nucleotides – Nucleotides contain: • Phosphate • Sugar, deoxyribose – Ribose in RNA • One of four nitrogenous bases (two purines and two pyrimidines)

5’-end 3’-end (free 3’-OH) 5’-end (free 5’phosphate) The two polynucleotide strands (the backbones) in the double helix run in opposite directions, and are said to be antiparallel Because of the pairing (A-T; G-C), one polynucleotide chain is always complementary to the base sequence of the other strand

Model questions

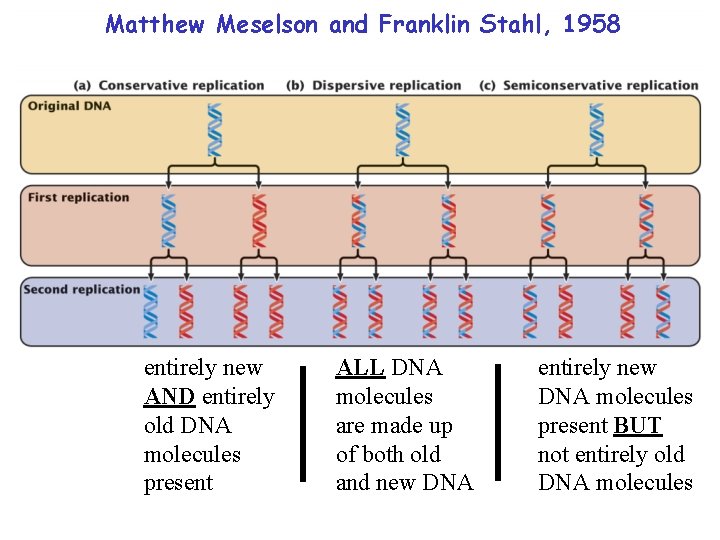
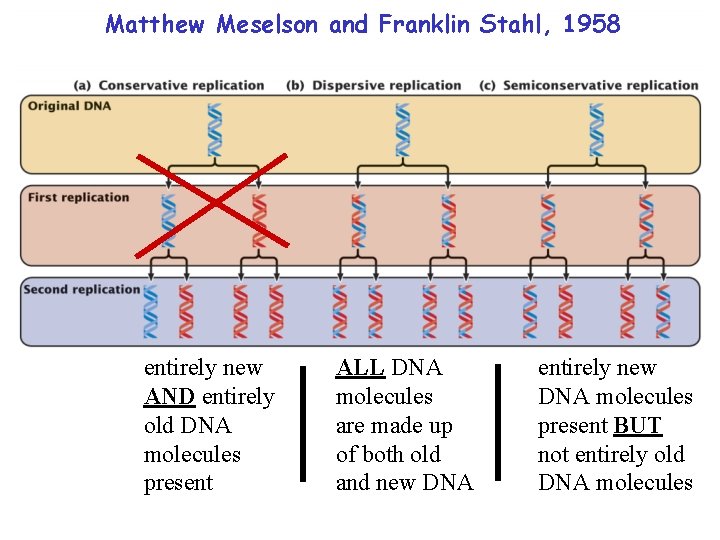
Matthew Meselson and Franklin Stahl, 1958 entirely new AND entirely old DNA molecules present ALL DNA molecules are made up of both old and new DNA entirely new DNA molecules present BUT not entirely old DNA molecules

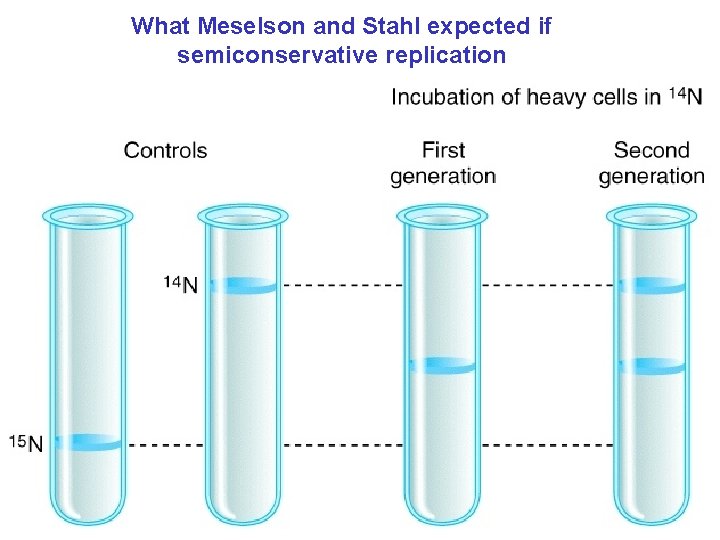
What Meselson and Stahl expected if semiconservative replication

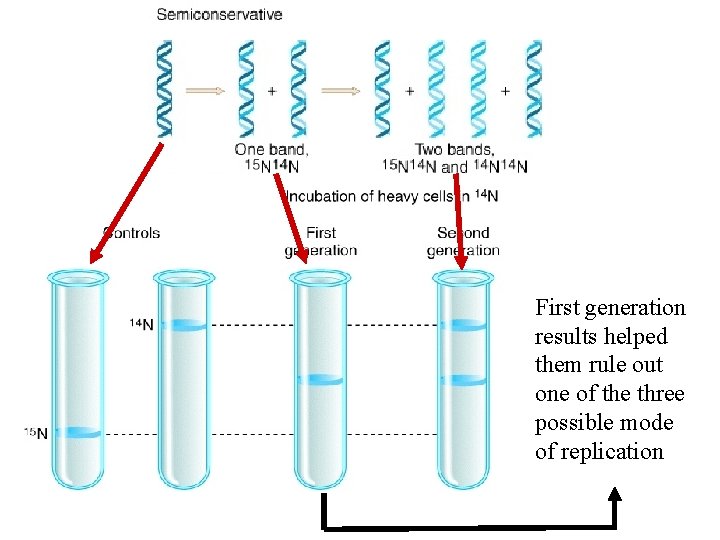
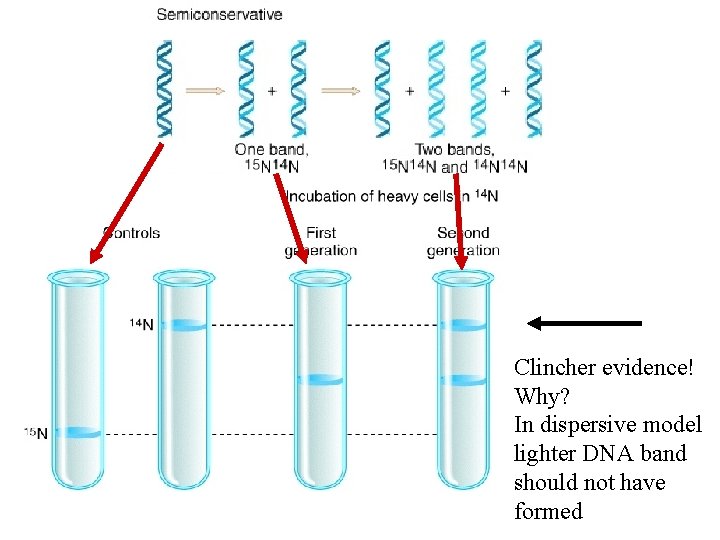
First generation results helped them rule out one of the three possible mode of replication

Matthew Meselson and Franklin Stahl, 1958 entirely new AND entirely old DNA molecules present ALL DNA molecules are made up of both old and new DNA entirely new DNA molecules present BUT not entirely old DNA molecules

Clincher evidence! Why? In dispersive model lighter DNA band should not have formed

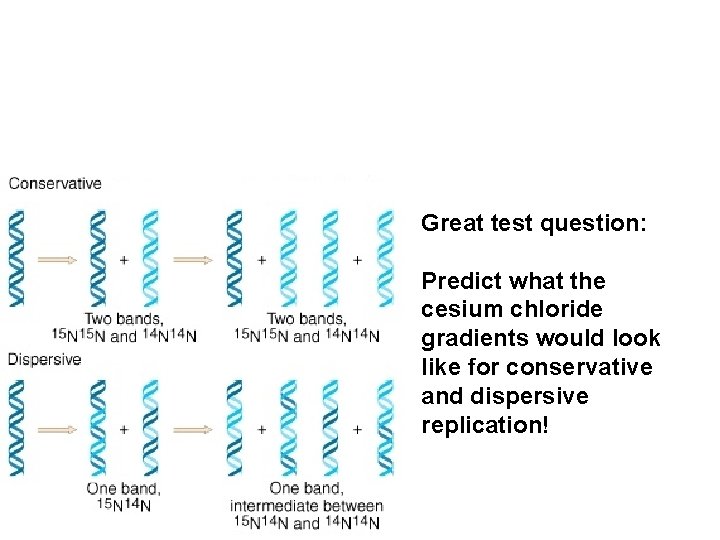
Great test question: Predict what the cesium chloride gradients would look like for conservative and dispersive replication!

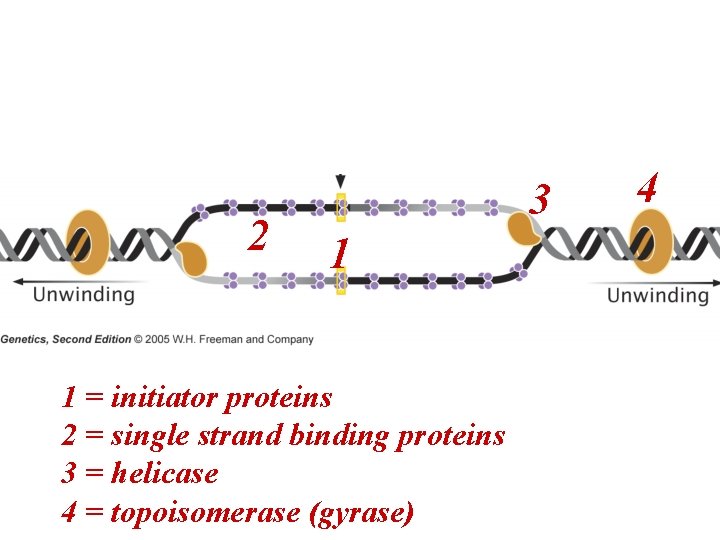
2 3 1 1 = initiator proteins 2 = single strand binding proteins 3 = helicase 4 = topoisomerase (gyrase) 4


Model questions

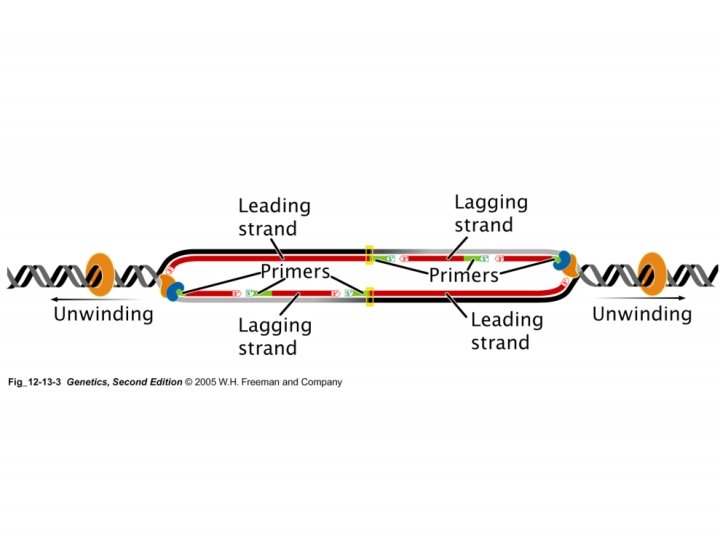
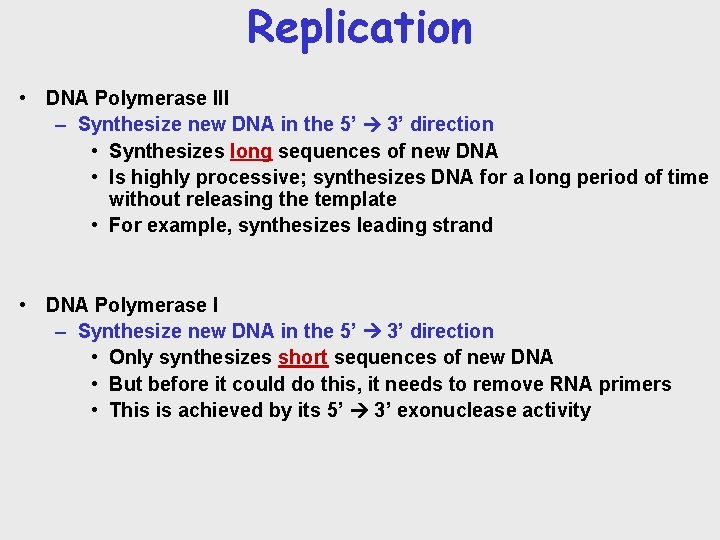
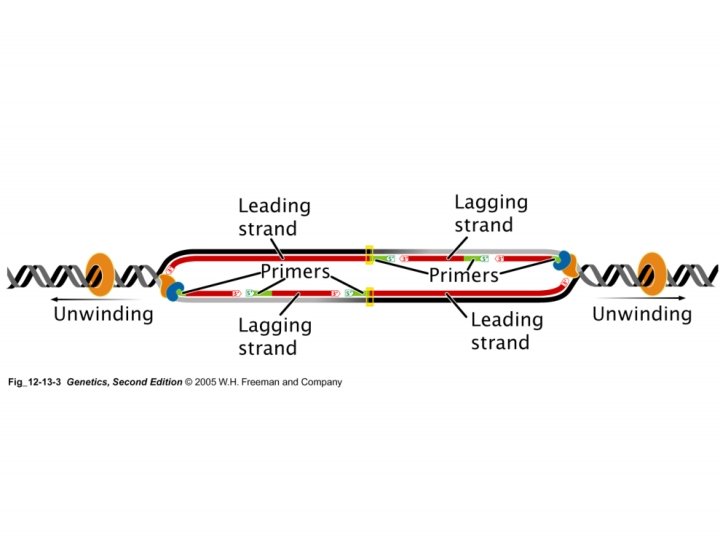
Replication • DNA Polymerase III – Synthesize new DNA in the 5’ 3’ direction • Synthesizes long sequences of new DNA • Is highly processive; synthesizes DNA for a long period of time without releasing the template • For example, synthesizes leading strand • DNA Polymerase I – Synthesize new DNA in the 5’ 3’ direction • Only synthesizes short sequences of new DNA • But before it could do this, it needs to remove RNA primers • This is achieved by its 5’ 3’ exonuclease activity

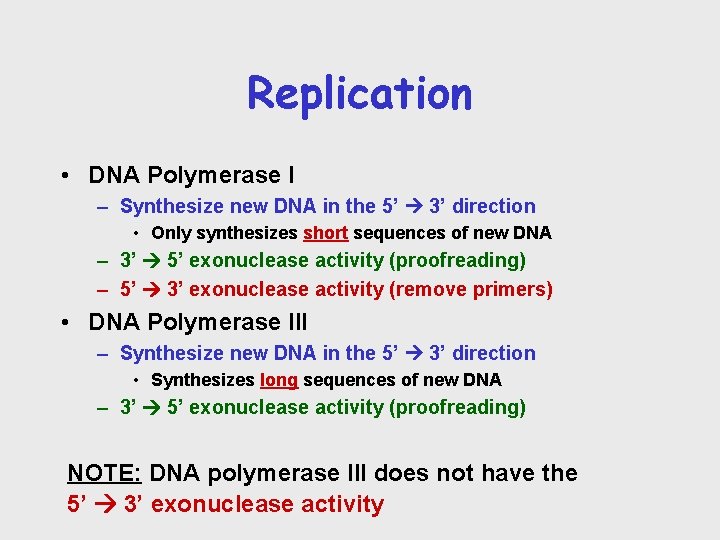
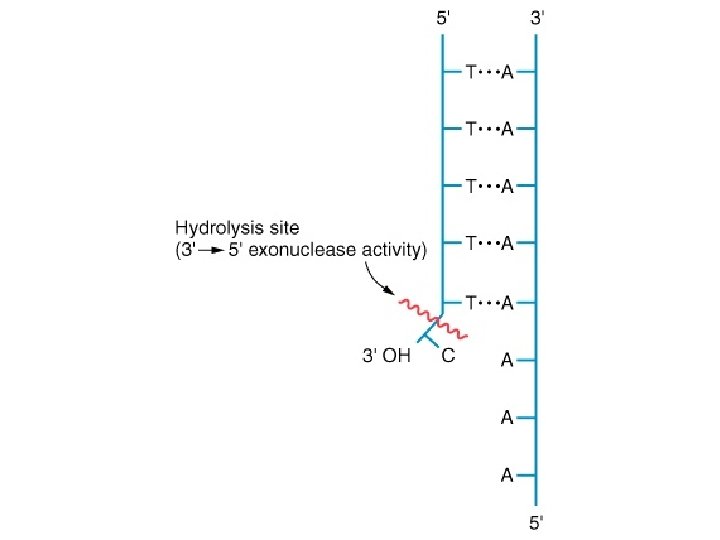
Replication • DNA Polymerase I – Synthesize new DNA in the 5’ 3’ direction • Only synthesizes short sequences of new DNA – 3’ 5’ exonuclease activity (proofreading) – 5’ 3’ exonuclease activity (remove primers) • DNA Polymerase III – Synthesize new DNA in the 5’ 3’ direction • Synthesizes long sequences of new DNA – 3’ 5’ exonuclease activity (proofreading) NOTE: DNA polymerase III does not have the 5’ 3’ exonuclease activity

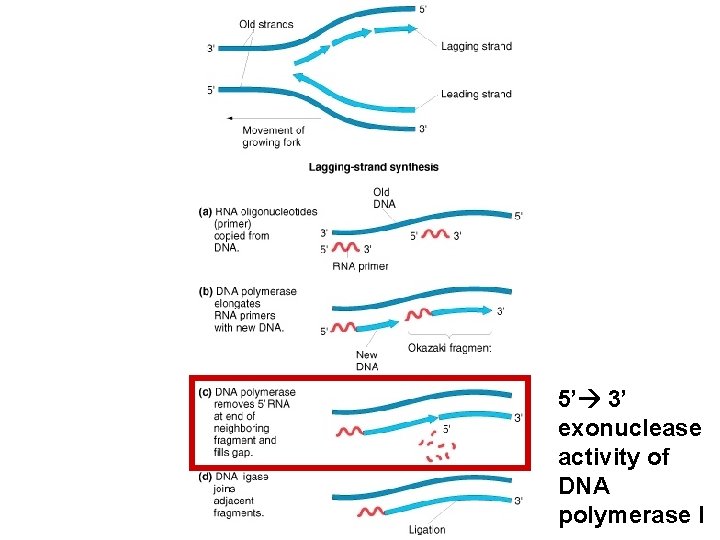
5’ 3’ exonuclease activity of DNA polymerase I


Model question You know that DNA ligase seals the gaps left behind by DNA polymerase I and completes DNA replication. You also know that both enzymes form the phosphodiester bonds. If that is the case why do cells need DNA ligase to seal the nicks and why not they use DNA polymerase I to seal these nicks?
