Molecular Genetics DNA Replication DNA Structure Nucleotides Deoxyribose
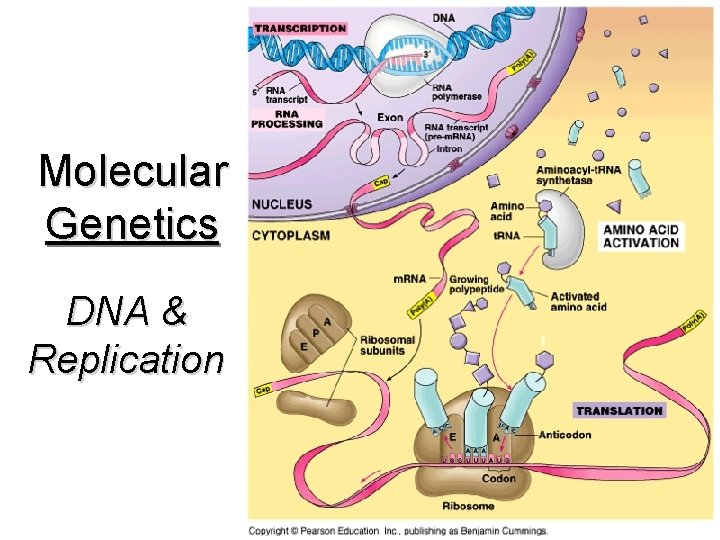
Molecular Genetics DNA & Replication
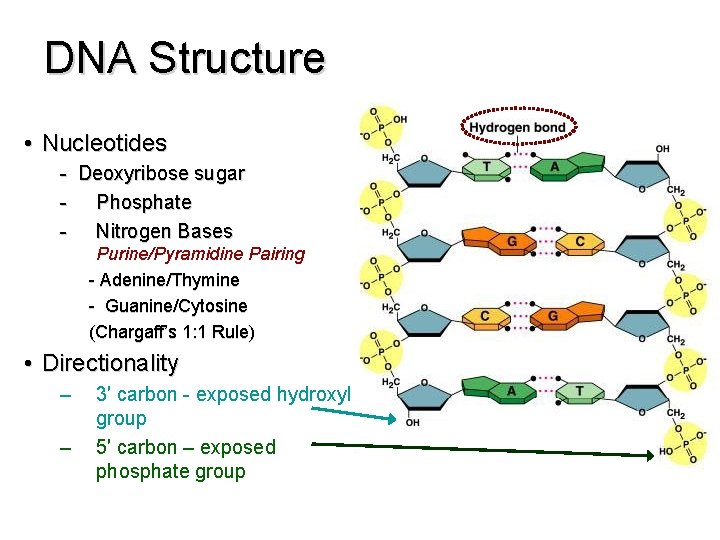
DNA Structure • Nucleotides - Deoxyribose sugar - Phosphate - Nitrogen Bases Purine/Pyramidine Pairing - Adenine/Thymine - Guanine/Cytosine (Chargaff’s 1: 1 Rule) • Directionality – – 3′ carbon - exposed hydroxyl group 5′ carbon – exposed phosphate group

History of DNA, Ch. 16 (know these for the exam): - Watson & Crick - Chargaff's rule - Franklin - Hershey & Chase - Willkins - Mc. Leod/Avery/Mc. Carty - Griffith
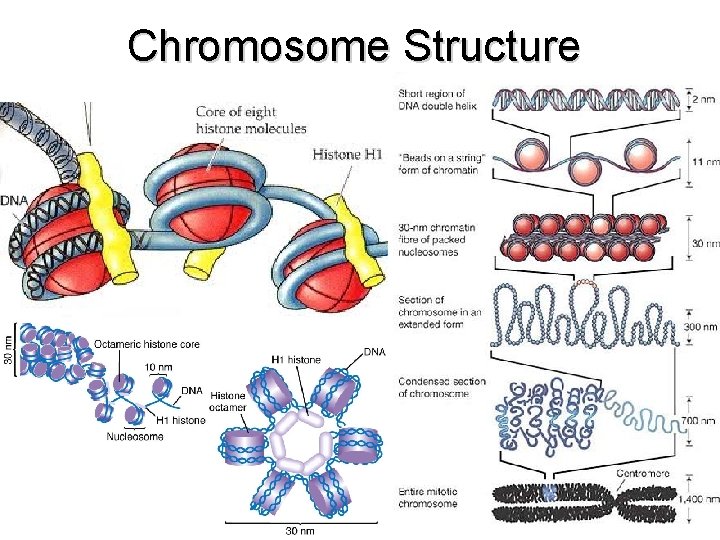
Chromosome Structure
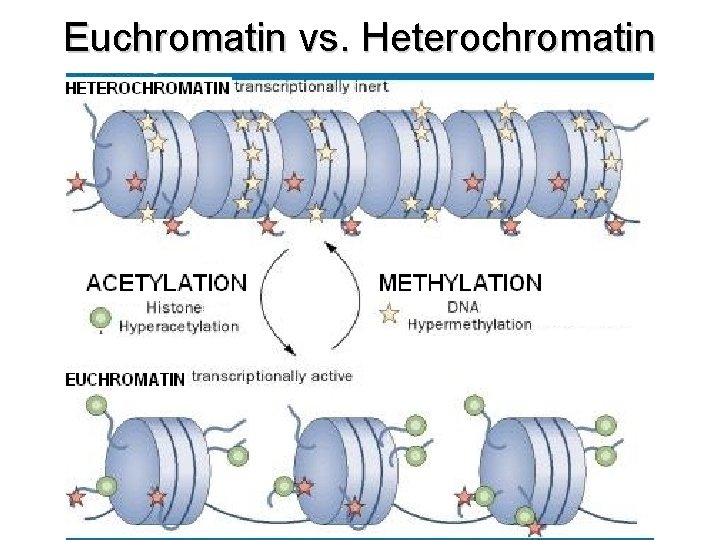
Euchromatin vs. Heterochromatin
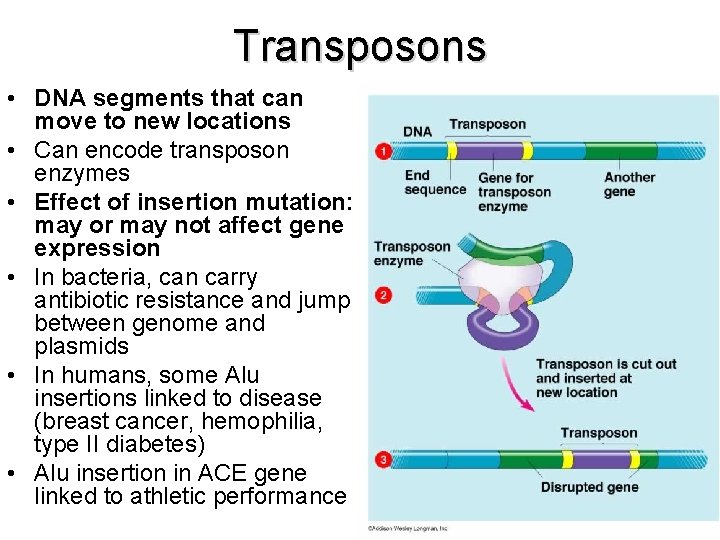
Transposons • DNA segments that can move to new locations • Can encode transposon enzymes • Effect of insertion mutation: may or may not affect gene expression • In bacteria, can carry antibiotic resistance and jump between genome and plasmids • In humans, some Alu insertions linked to disease (breast cancer, hemophilia, type II diabetes) • Alu insertion in ACE gene linked to athletic performance

Chromosome Structure • http: //www. youtube. com/watch? v=q. Ys. W 0 j. I FH 5 A (1: 17) • http: //www. youtube. com/watch? v=e. Yr. Q 0 E h. VCYA (2: 18) • http: //www. youtube. com/watch? v=gb. SIBh Fw. Q 4 s (1: 42)

DNA Replication • Semiconservative • ½ old – template strand • ½ new – complementary strand • Requires enzymes • Copy: Antiparallel 5′ → 3′ direction Leading Strand Lagging Strand
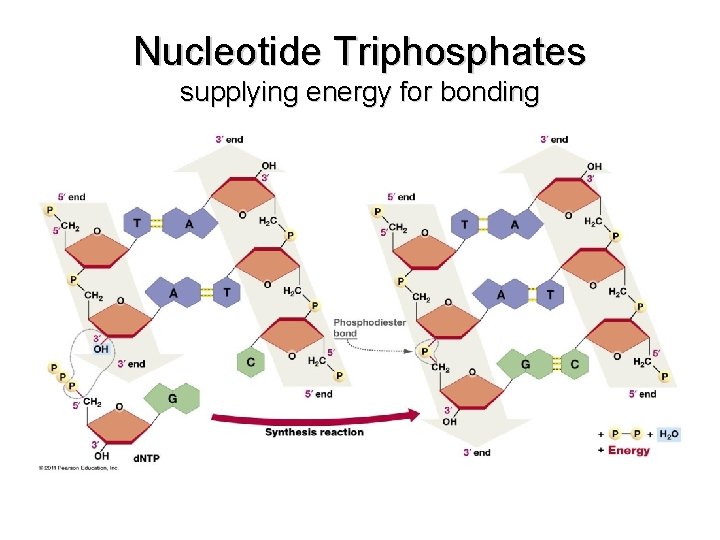
Nucleotide Triphosphates supplying energy for bonding
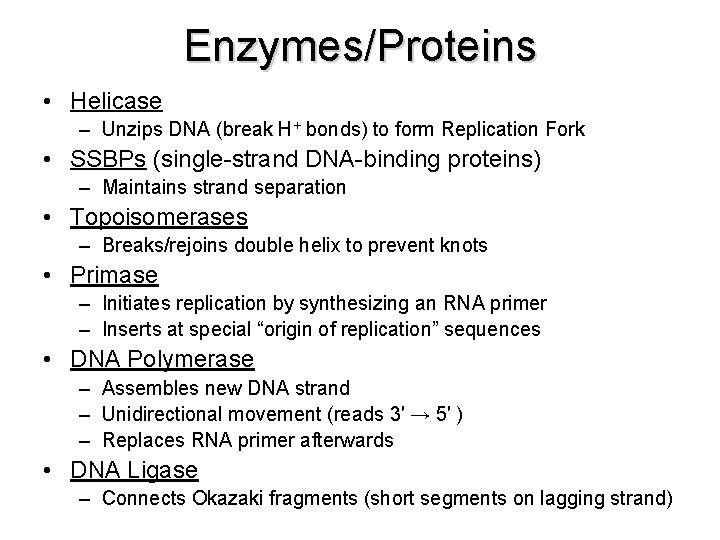
Enzymes/Proteins • Helicase – Unzips DNA (break H+ bonds) to form Replication Fork • SSBPs (single-strand DNA-binding proteins) – Maintains strand separation • Topoisomerases – Breaks/rejoins double helix to prevent knots • Primase – Initiates replication by synthesizing an RNA primer – Inserts at special “origin of replication” sequences • DNA Polymerase – Assembles new DNA strand – Unidirectional movement (reads 3′ → 5′ ) – Replaces RNA primer afterwards • DNA Ligase – Connects Okazaki fragments (short segments on lagging strand)
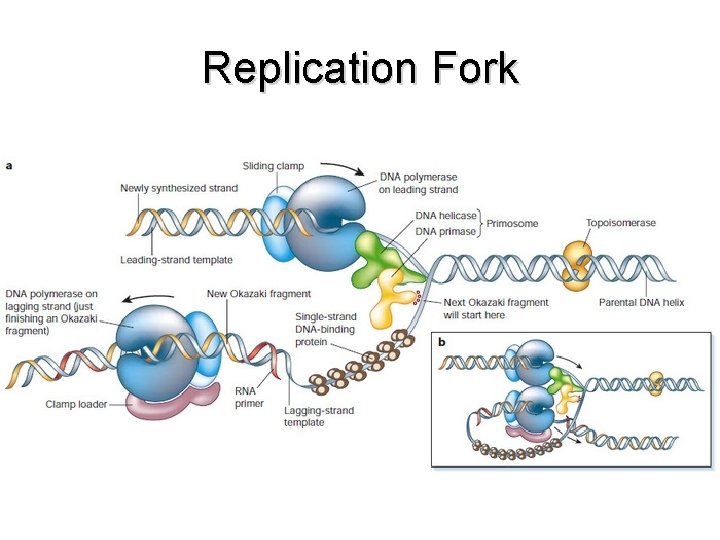
Replication Fork
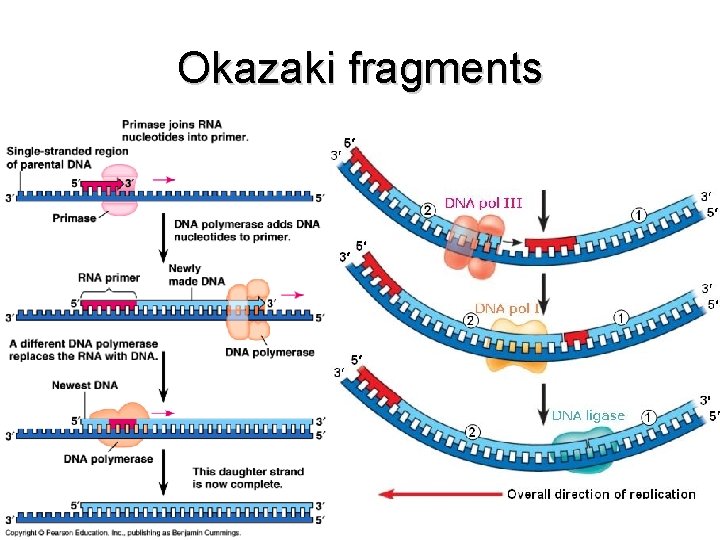
Okazaki fragments

DNA Replication Animations • http: //www. youtube. com/watch? v=mt. LXpgj. HL 0 (2: 04) • http: //www. youtube. com/watch? v=I 9 Ar. IJW YZHI (2: 18)
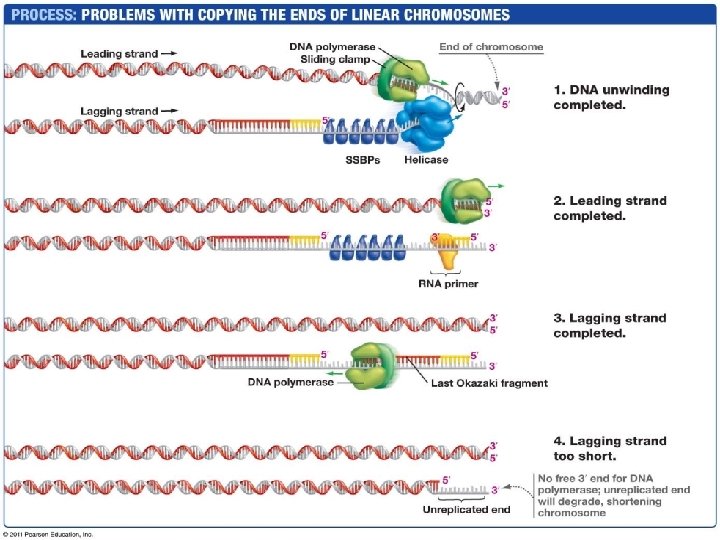

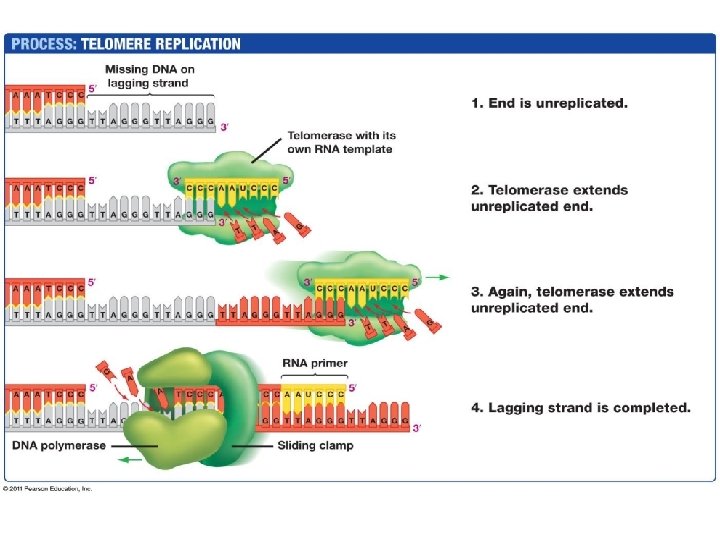
Telomeres • The ends of eukaryotic (linear) chromosomes – Repeating nucleotides (vertebrates: TTAGGG x 1000+) • Lagging strand has no primer at one end, leaving a singlestranded section of template that will be degraded (shortening the chromosome) • Telomerase adds more repeating bases to the lagging strand – Prevents the lagging strand from getting shorter with each replication – Absent in somatic cells: chromosomes get progressively shorter over time, limiting lifespan of tissue – Secret to immortality? – Cancer cells express telomerase

Telomerase Animations • http: //www. youtube. com/watch? v=AJNo. T m. Ws. E 0 s (2: 06) • http: //www. youtube. com/watch? v=DV 3 Xjq W_xg. U (4: 38) *if time
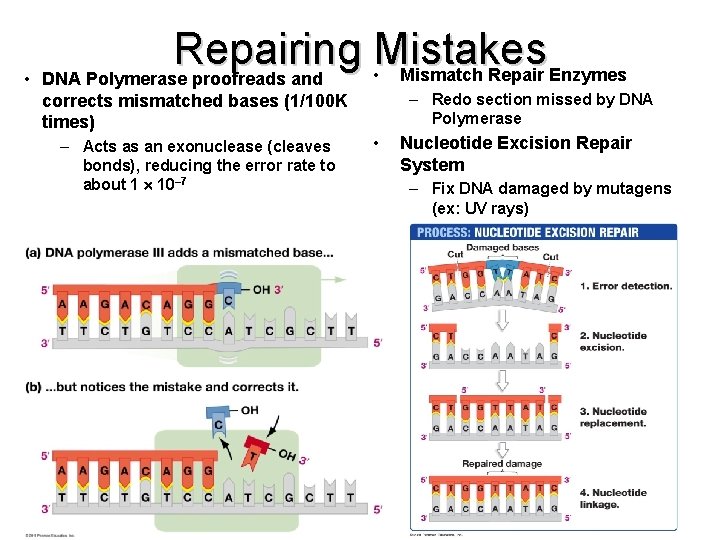
Repairing Mistakes • Mismatch Repair Enzymes • DNA Polymerase proofreads and – Redo section missed by DNA Polymerase corrects mismatched bases (1/100 K times) – Acts as an exonuclease (cleaves bonds), reducing the error rate to about 1 10– 7 • Nucleotide Excision Repair System – Fix DNA damaged by mutagens (ex: UV rays)

Nucleotide Excision Repair • http: //www. youtube. com/watch? v=u. N 82 GL QYAUQ (1: 05) • http: //www. youtube. com/watch? v=Cc. Tayx Eblio (0: 42) • http: //www. youtube. com/watch? v=bg. UH 9 Nf. O 2 QM (0: 27)
- Slides: 19