Molecular dynamics study of gas phase and gassurface










- Slides: 55

Molecular dynamics study of gas phase and gas-surface reaction using “MD Trajectory” software complex Michael Pogosbekian Valery Kovalev Institute of Mechanics, Moscow State University Dept. Mechanics and Mathematics, Moscow State University

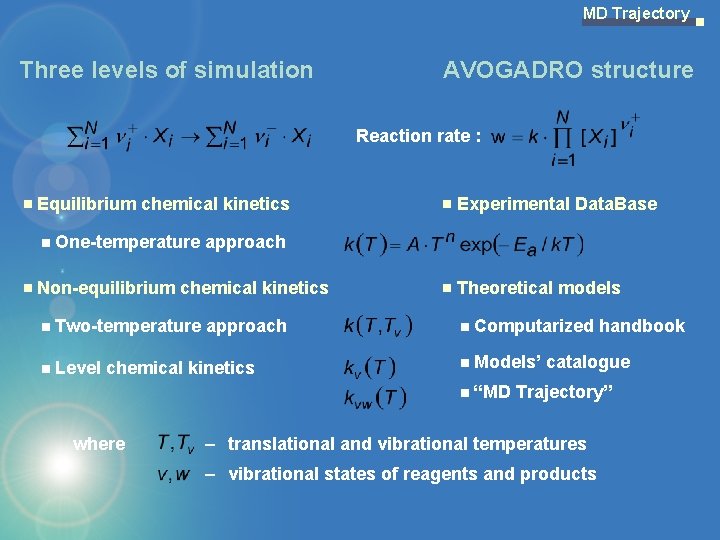
MD Trajectory Three levels of simulation AVOGADRO structure Reaction rate : Equilibrium chemical kinetics Experimental Data. Base One-temperature approach Non-equilibrium chemical kinetics Theoretical models Two-temperature approach Computarized handbook Level chemical kinetics Models’ catalogue “MD Trajectory” where – translational and vibrational temperatures – vibrational states of reagents and products

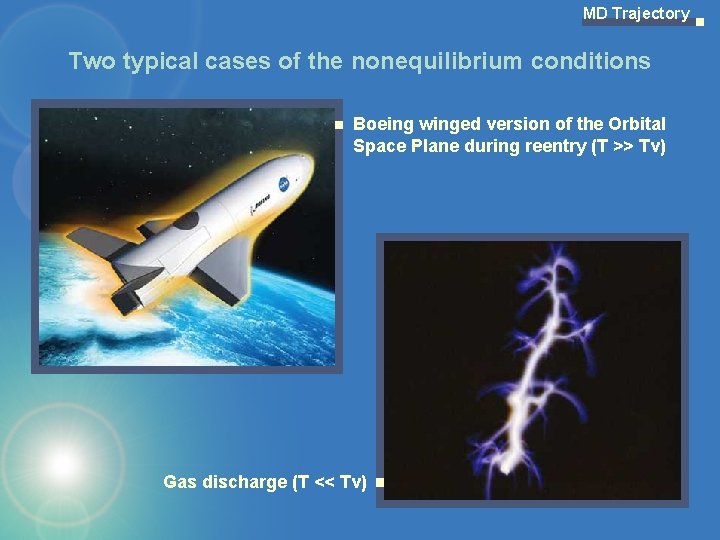
MD Trajectory Two typical cases of the nonequilibrium conditions Boeing winged version of the Orbital Space Plane during reentry (T >> Tv) Gas discharge (T << Tv)

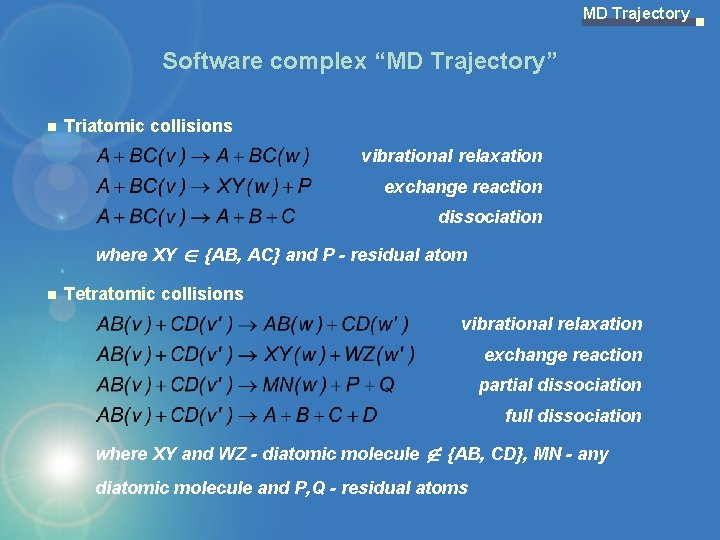
MD Trajectory Software complex “MD Trajectory” Triatomic collisions vibrational relaxation exchange reaction dissociation where XY Î {AB, AC} and P - residual atom Tetratomic collisions vibrational relaxation exchange reaction partial dissociation full dissociation where XY and WZ - diatomic molecule Ï {AB, CD}, MN - any diatomic molecule and P, Q - residual atoms

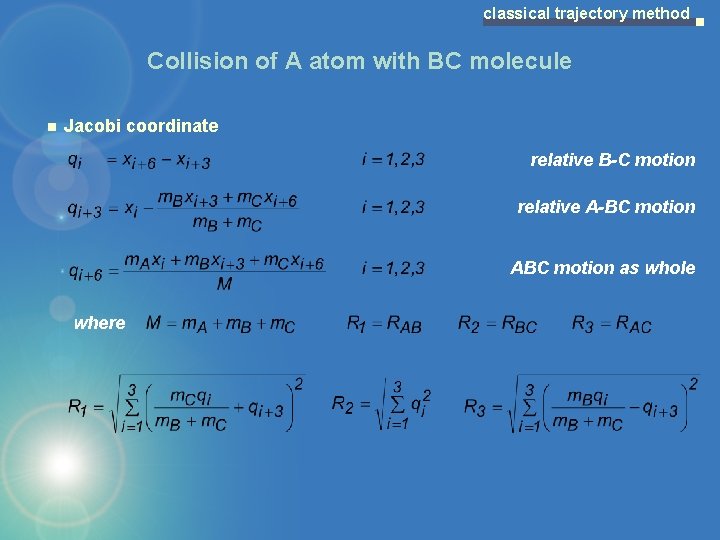
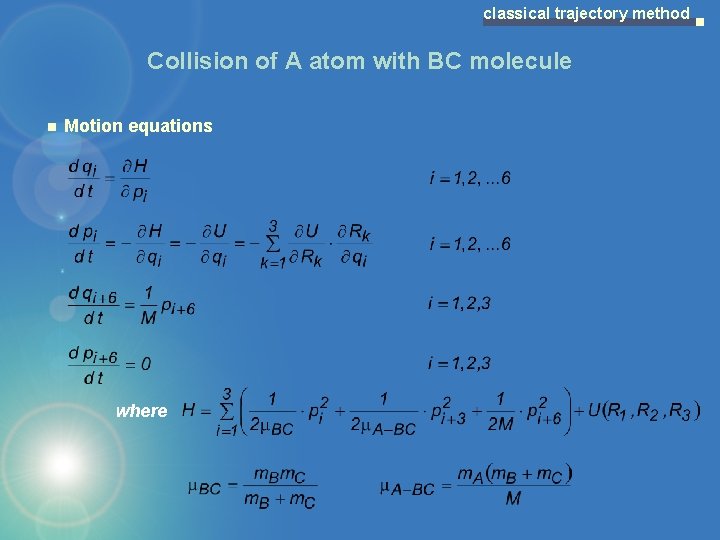
classical trajectory method Collision of A atom with BC molecule Jacobi coordinate relative B-C motion relative A-BC motion ABC motion as whole where

classical trajectory method Collision of A atom with BC molecule Motion equations where

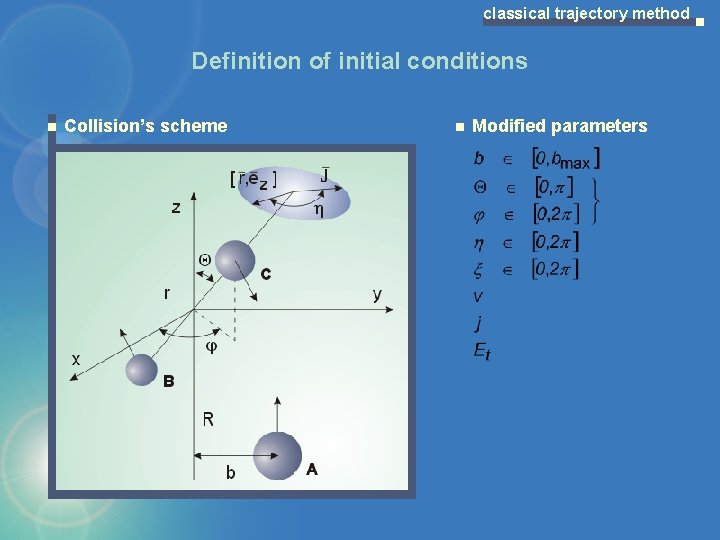
classical trajectory method Definition of initial conditions Collision’s scheme Modified parameters

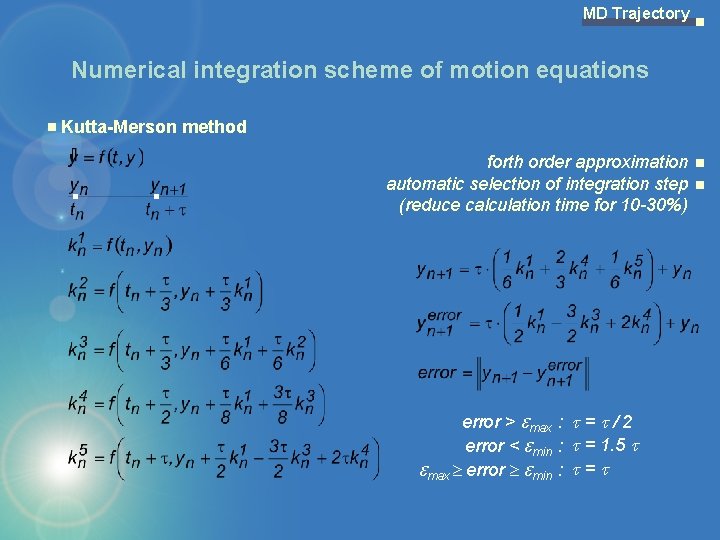
MD Trajectory Numerical integration scheme of motion equations Kutta-Merson method forth order approximation automatic selection of integration step (reduce calculation time for 10 -30%) error > max : = / 2 error < min : = 1. 5 max error min : =

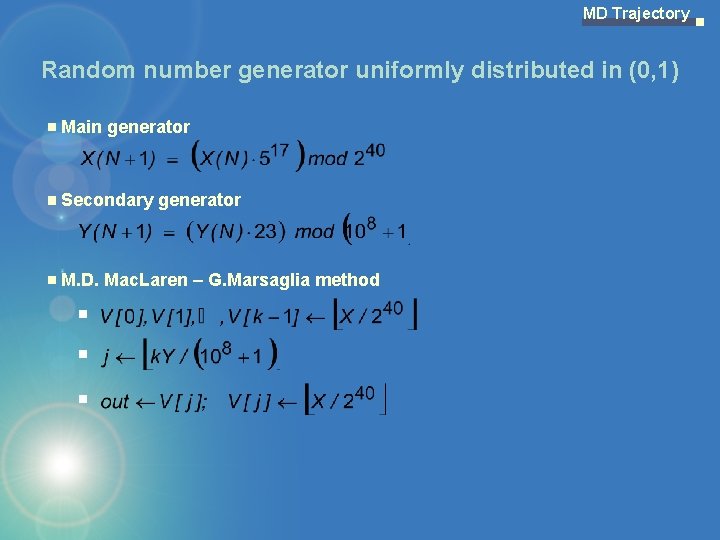
MD Trajectory Random number generator uniformly distributed in (0, 1) Main generator Secondary generator M. D. Mac. Laren – G. Marsaglia method

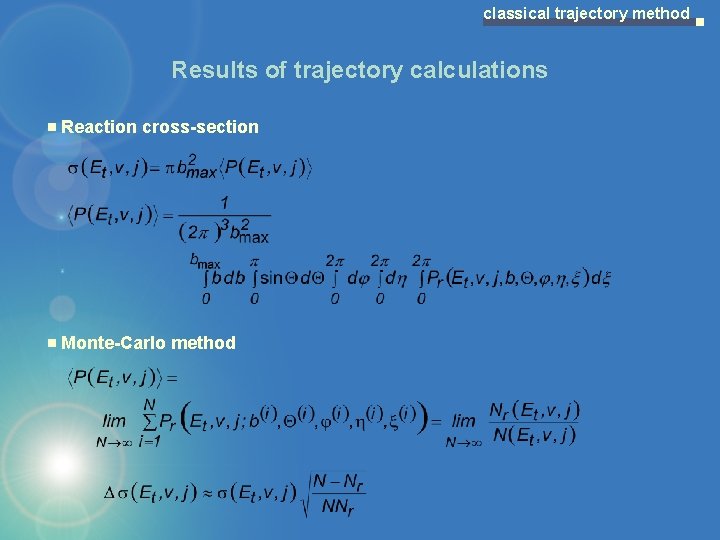
classical trajectory method Results of trajectory calculations Reaction cross-section Monte-Carlo method

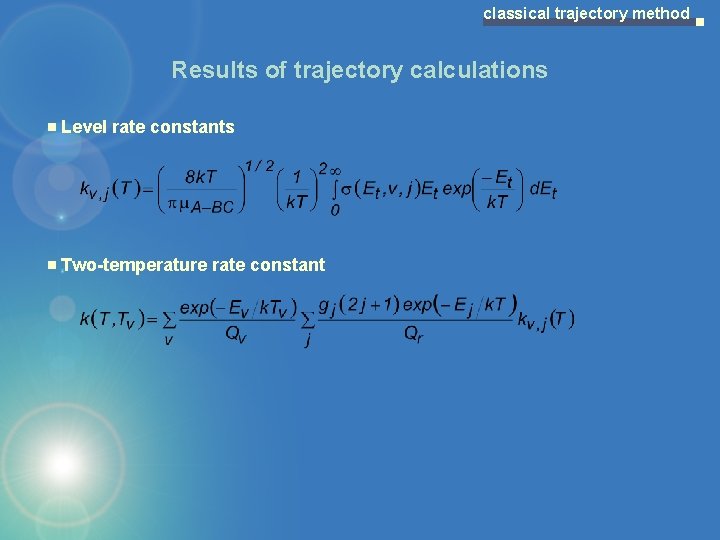
classical trajectory method Results of trajectory calculations Level rate constants Two-temperature rate constant

PES Potential energy surface Semiempirical methods Generalized LEPS model Method of diatomic complexes in molecules Bond energy- bond order method Ab-initio calculations GAUSSIAN MOLCAS GAMESS (special version for INTEL platform - PC GAMESS)

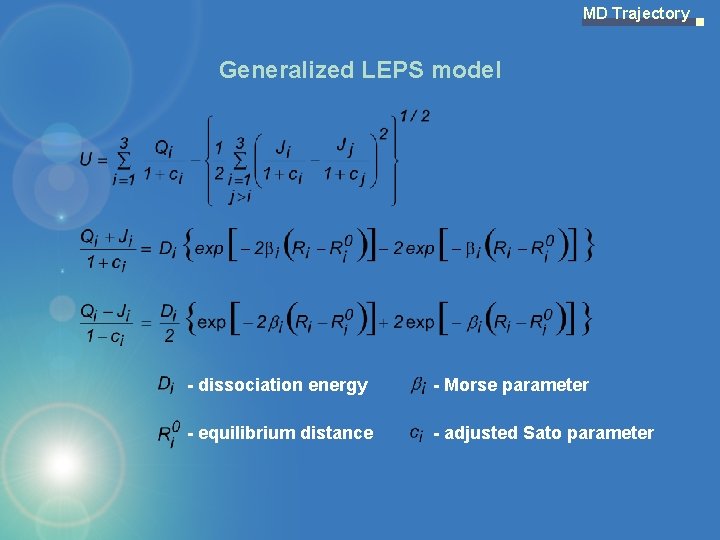
MD Trajectory Generalized LEPS model - dissociation energy - Morse parameter - equilibrium distance - adjusted Sato parameter

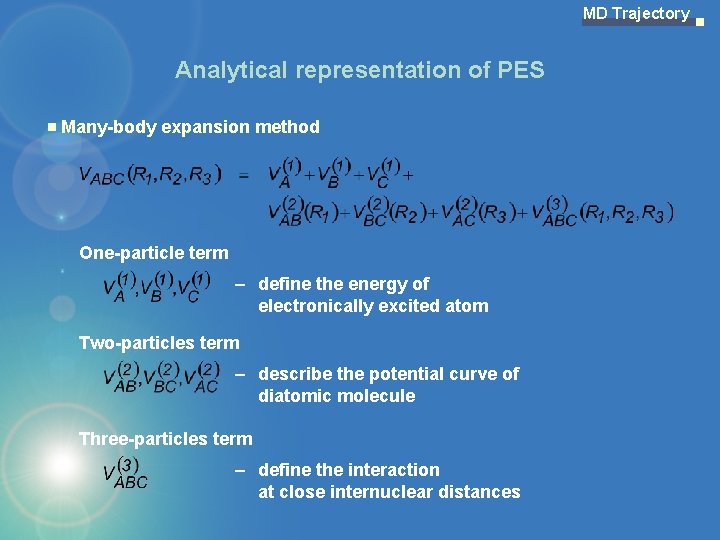
MD Trajectory Analytical representation of PES Many-body expansion method One-particle term – define the energy of electronically excited atom Two-particles term – describe the potential curve of diatomic molecule Three-particles term – define the interaction at close internuclear distances

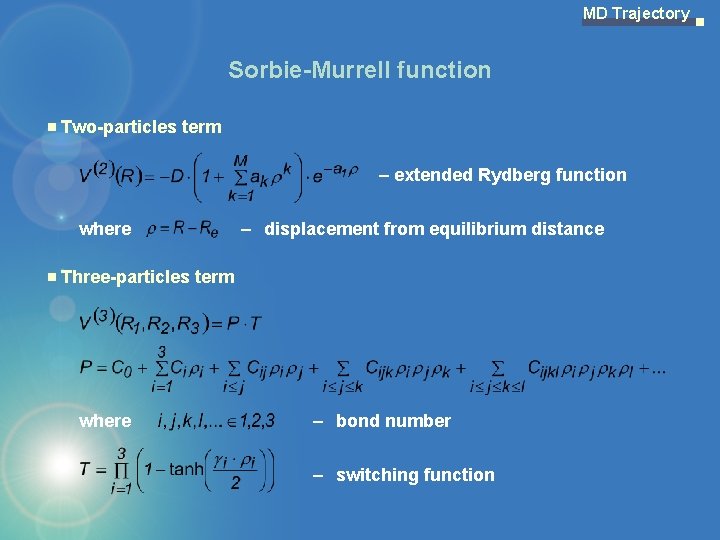
MD Trajectory Sorbie-Murrell function Two-particles term – extended Rydberg function where – displacement from equilibrium distance Three-particles term where – bond number – switching function

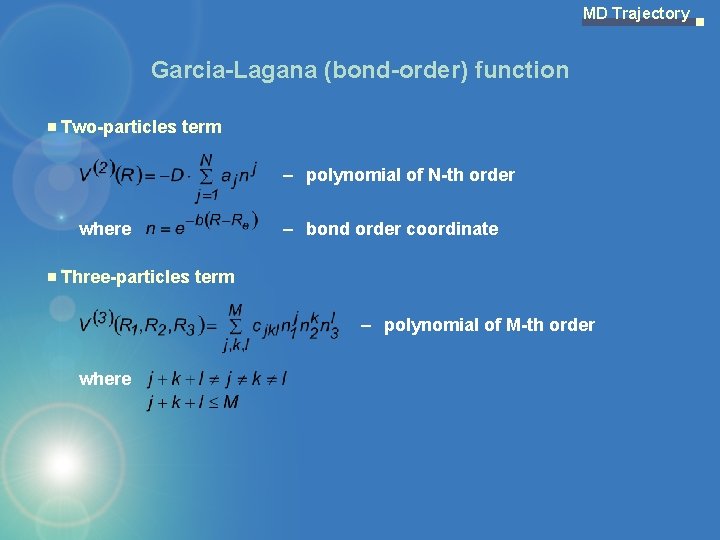
MD Trajectory Garcia-Lagana (bond-order) function Two-particles term – polynomial of N-th order where – bond order coordinate Three-particles term – polynomial of M-th order where

MD Trajectory Aguado-Paniagua function Two-particles term where l = 2 or 3 for two- or three-particles term Three-particles term – polynomial of M-th order where

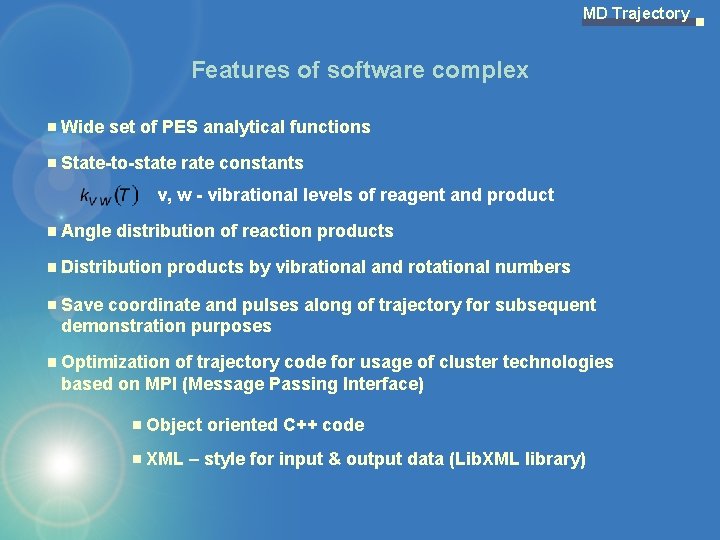
MD Trajectory Features of software complex Wide set of PES analytical functions State-to-state rate constants v, w - vibrational levels of reagent and product Angle distribution of reaction products Distribution products by vibrational and rotational numbers Save coordinate and pulses along of trajectory for subsequent demonstration purposes Optimization of trajectory code for usage of cluster technologies based on MPI (Message Passing Interface) Object oriented C++ code XML – style for input & output data (Lib. XML library)

MD Trajectory High performance supercomputer facilities Moscow State University, cluster SCI - 36 CPUs total Node configuration: Dual Pentium III/500 MHz, 1 Gb RAM, 3. 2 Gb HDD Network environment : SCI + Fast Ethernet Russian Academy of Sciences, cluster MVS-1000 M - 768 CPUs total Node configuration: Dual Alpha 21264 A/667 MHz, 1 Gb RAM, 15 Gb HDD Network environment : Myrinet (2 Gb/s) + Fast Ethernet

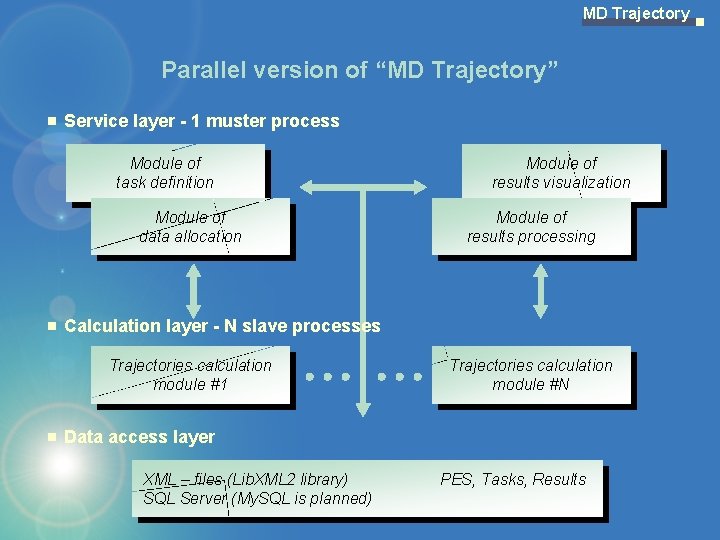
MD Trajectory Parallel version of “MD Trajectory” Service layer - 1 muster process Module of task definition Module of data allocation Module of results visualization Module of results processing Calculation layer - N slave processes Trajectories calculation module #1 Trajectories calculation module #N Data access layer XML – files (Lib. XML 2 library) SQL Server (My. SQL is planned) PES, Tasks, Results

MD Trajectory Data Access Layer Three kinds of input & output files Input data – fixed size – rather simple structure – less than 1 Mb Control Group describes control function; Molecule Group contains spectroscopic characteristic of diatomic molecules PES Group describes PES of the investigated system Log file – dynamic size – rather simple structure – less than 10 Mb Auxiliary information which can be required for calculation control and calculation continuation at the next time Result file – dynamic size – very complex structure – up to 100 Mb Two ways for realization of Data Access Layer XML files. It is realized due to Lib. XML 2 library XML parser and toolkit of Gnome WWW. XMLSOFT. ORG Data. Base connectivity module for My. SQL Server is planned

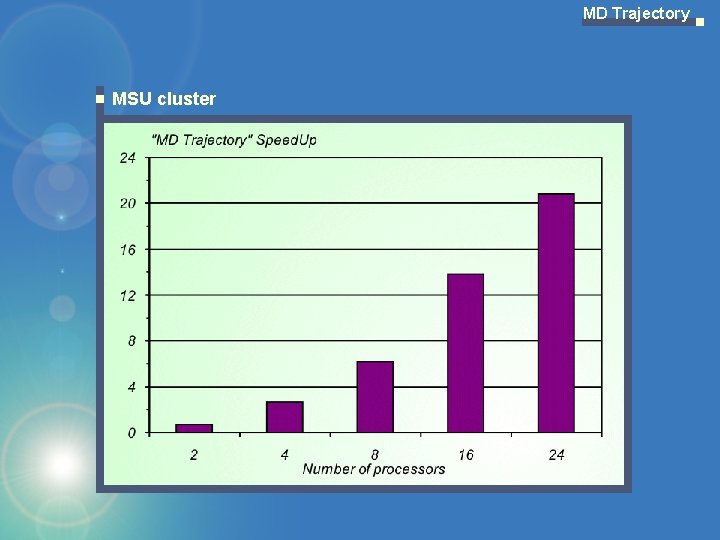
MD Trajectory MSU cluster

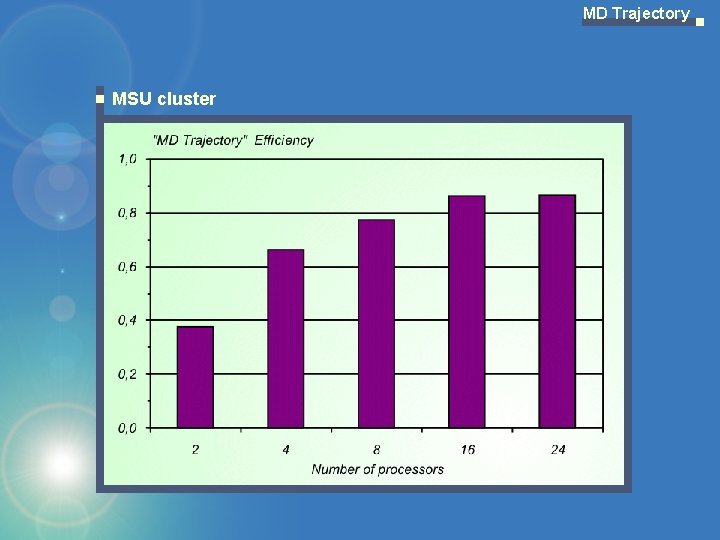
MD Trajectory MSU cluster

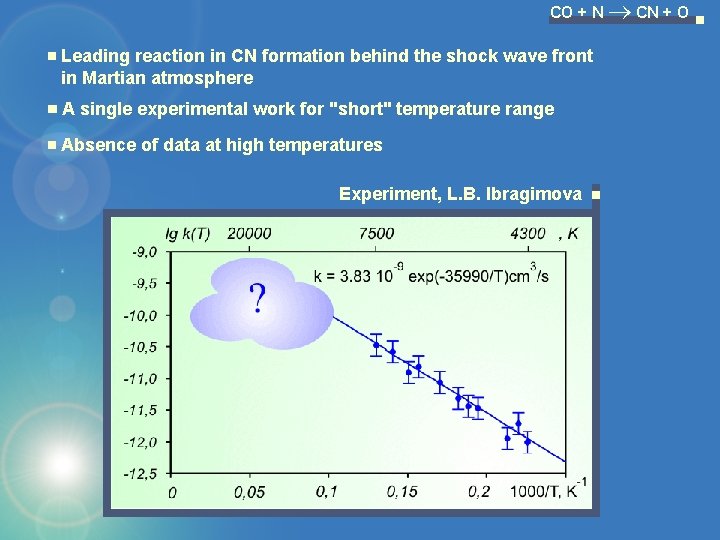
CO + N CN + O Leading reaction in CN formation behind the shock wave front in Martian atmosphere A single experimental work for "short" temperature range Absence of data at high temperatures Experiment, L. B. Ibragimova

CO + N CN + O Potential energy surface PES for Modified LEPS model [1] Sorbie-Murrell function [3] Aguado-Paniagua function [4] based on ab-initio data [2, 4] PES for Generalized LEPS model [1] References 1. K. J. Schmatjko and J. Wolfrum, Ber. Bunsen Phys. Chem. , 1975, 79, pp. 696 -707 2. P. Halvick, J. C. Rayez, E. M. Evleth, J. Chem. Phys. , 1984, 81, pp. 728 -737 3. SM. Simonson, N. Markovic, S. Nordholm and B. J. Persson, Chem. Phys. , 1995, 200, pp. 141 -160 4. Andersson, N. Markovic and G. Nyman, Phys. Chem. Phys. , 2000, 2, pp. 613 -620

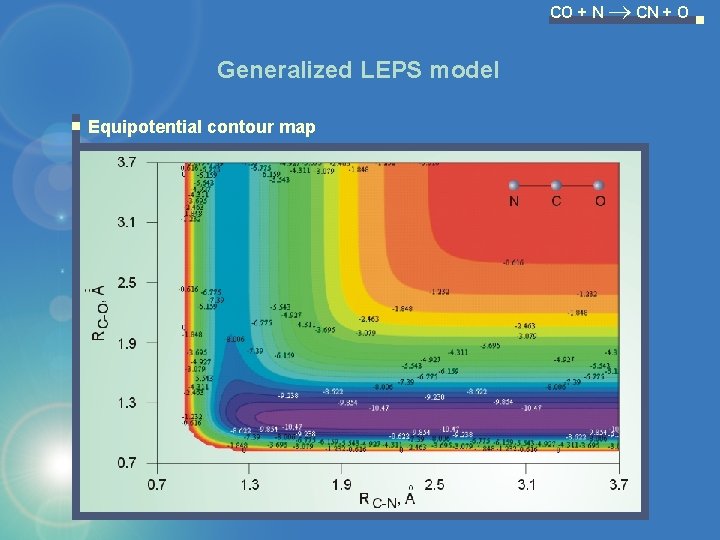
CO + N CN + O Generalized LEPS model Equipotential contour map

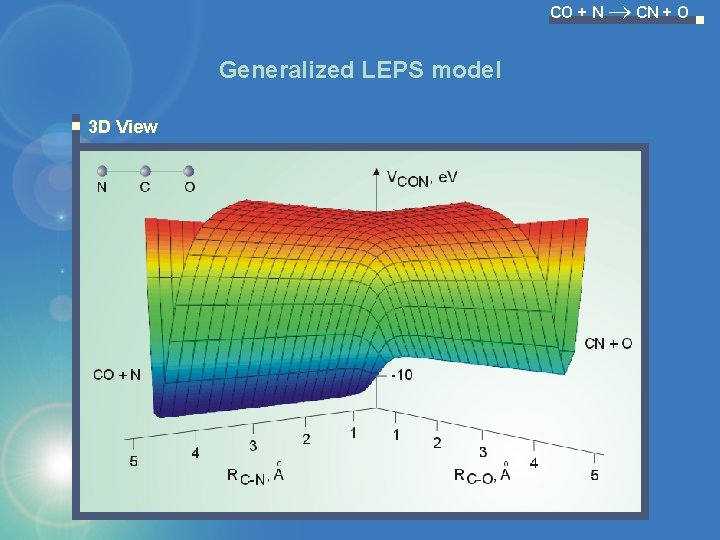
CO + N CN + O Generalized LEPS model 3 D View

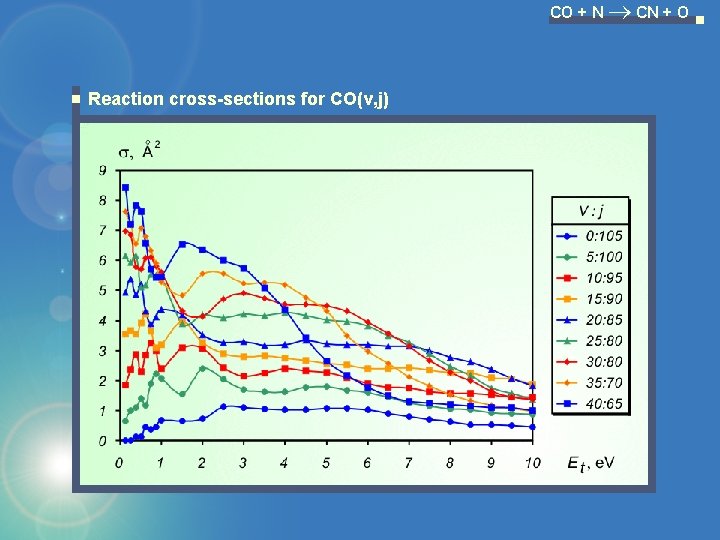
CO + N CN + O Reaction cross-sections for CO(v, j)

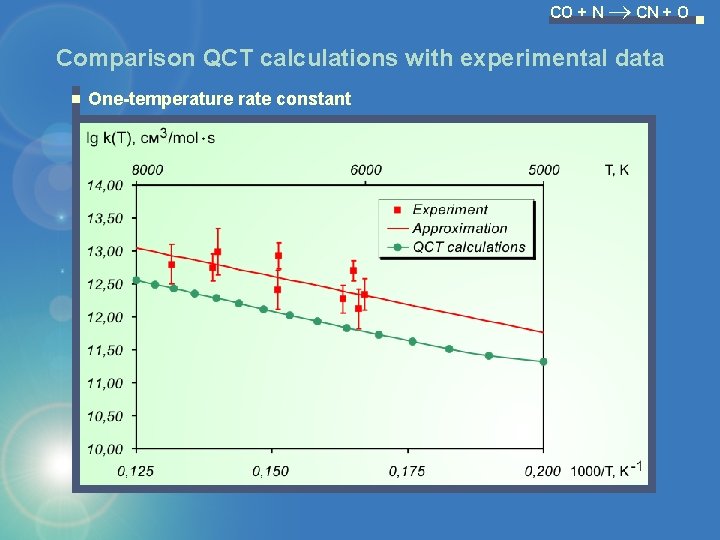
CO + N CN + O Comparison QCT calculations with experimental data One-temperature rate constant

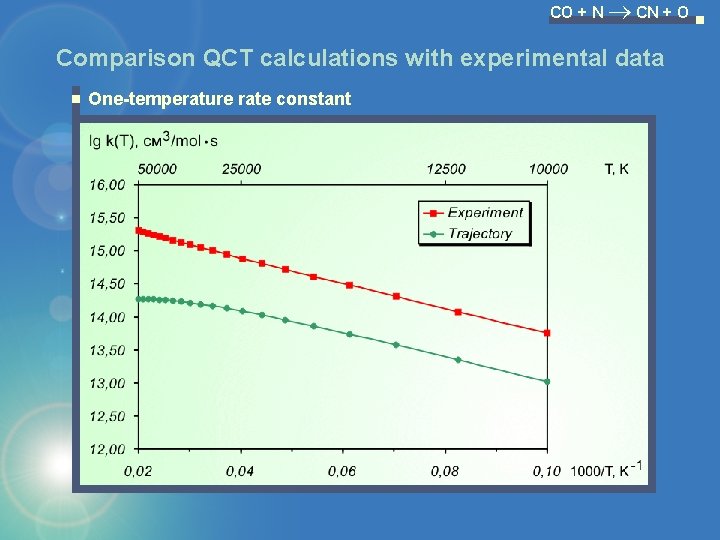
CO + N CN + O Comparison QCT calculations with experimental data One-temperature rate constant

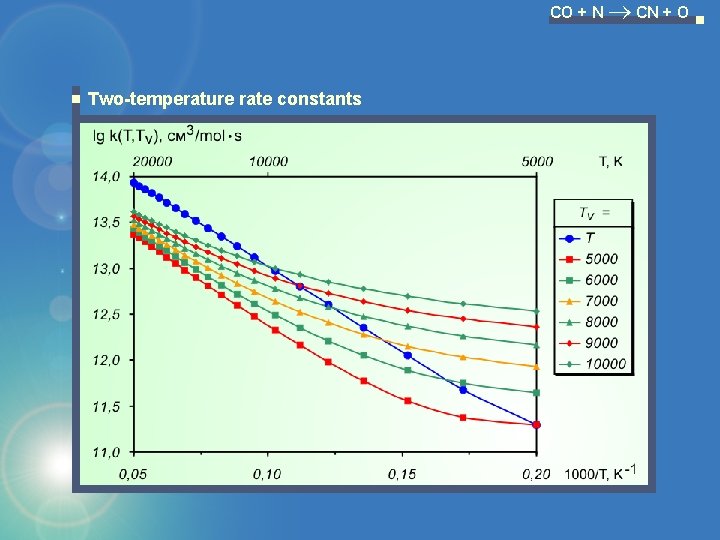
CO + N CN + O Two-temperature rate constants

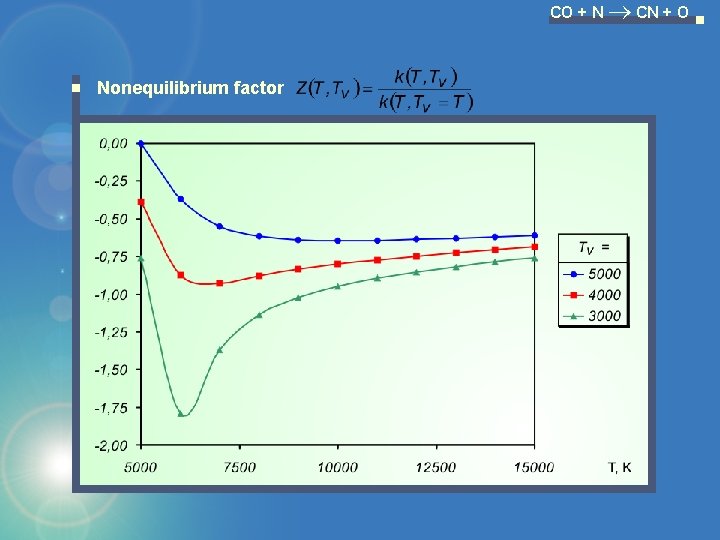
CO + N CN + O Nonequilibrium factor

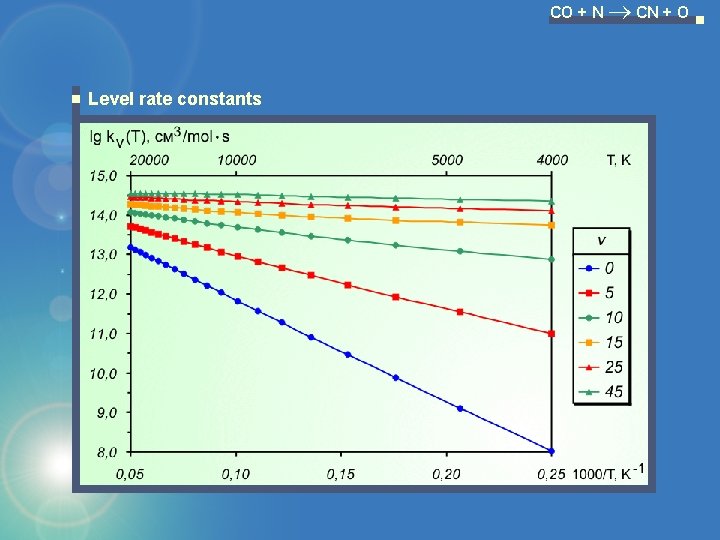
CO + N CN + O Level rate constants

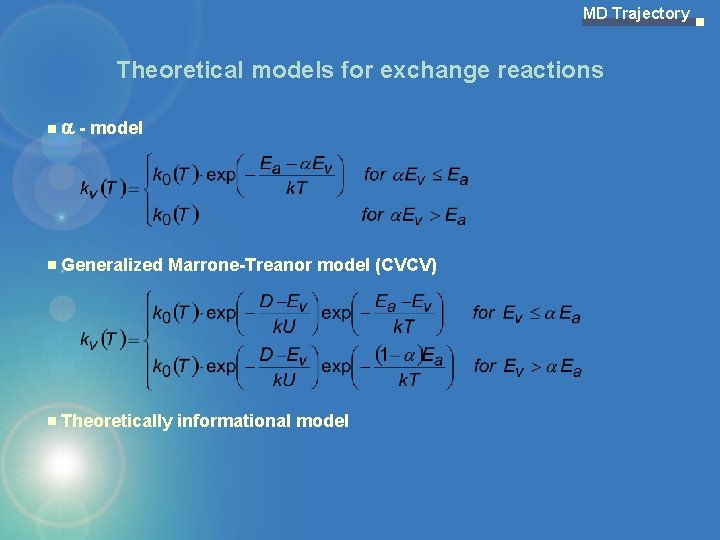
MD Trajectory Theoretical models for exchange reactions - model Generalized Marrone-Treanor model (CVCV) Theoretically informational model

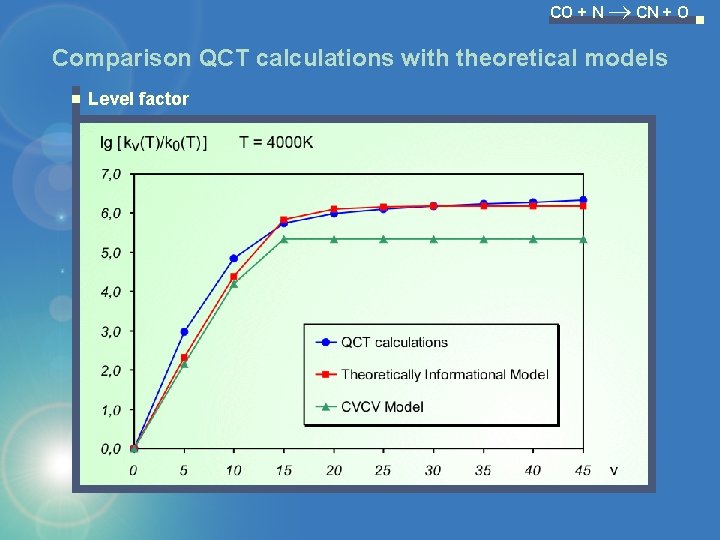
CO + N CN + O Comparison QCT calculations with theoretical models Level factor

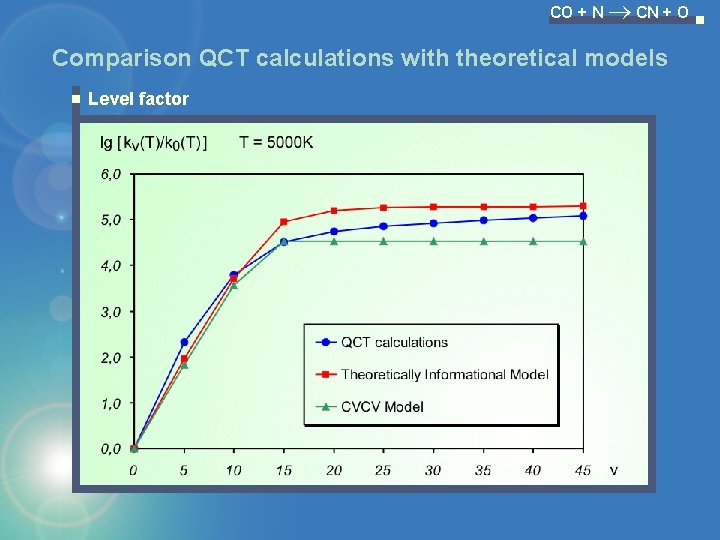
CO + N CN + O Comparison QCT calculations with theoretical models Level factor

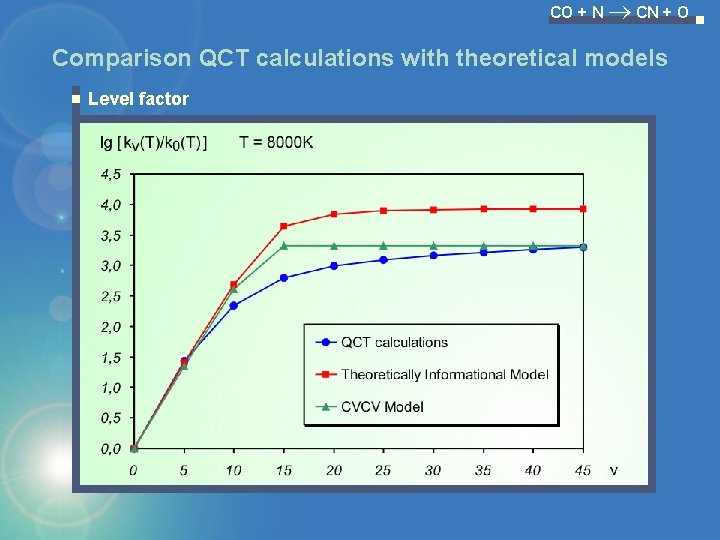
CO + N CN + O Comparison QCT calculations with theoretical models Level factor

CO + N CN + O Comparison QCT calculations with theoretical models Level factor

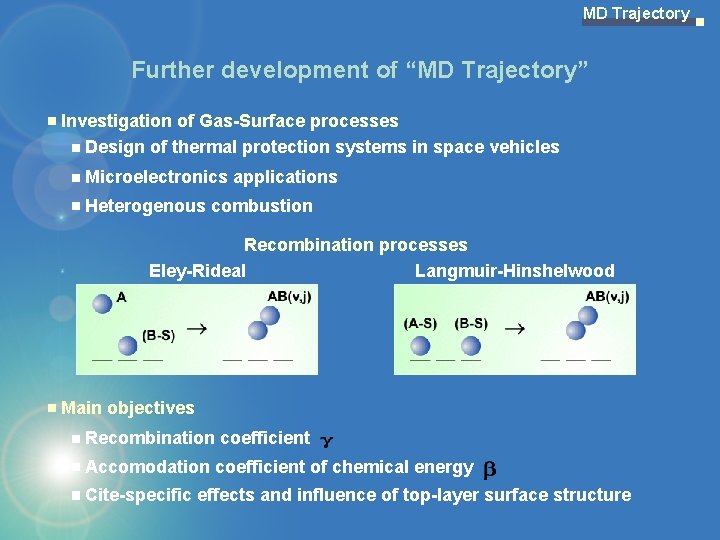
MD Trajectory Further development of “MD Trajectory” Investigation of Gas-Surface processes Design of thermal protection systems in space vehicles Microelectronics applications Heterogenous combustion Recombination processes Eley-Rideal Langmuir-Hinshelwood Main objectives Recombination coefficient Accomodation coefficient of chemical energy Cite-specific effects and influence of top-layer surface structure

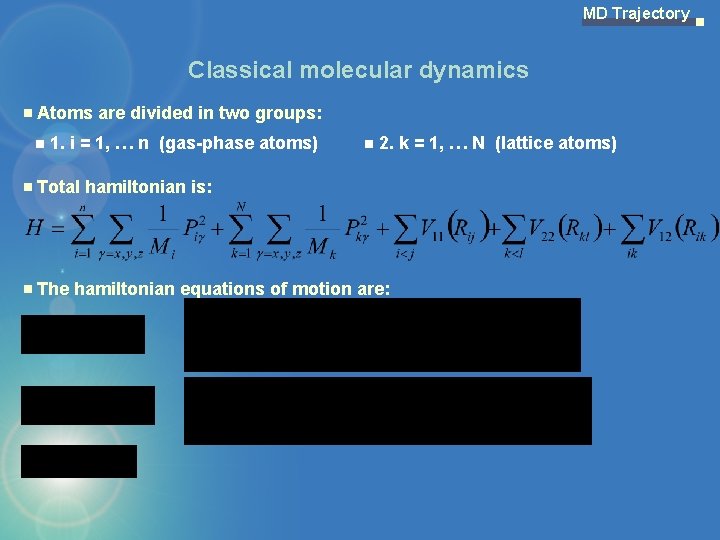
MD Trajectory Classical molecular dynamics Atoms are divided in two groups: 1. i = 1, … n (gas-phase atoms) 2. k = 1, … N (lattice atoms) Total hamiltonian is: The hamiltonian equations of motion are:

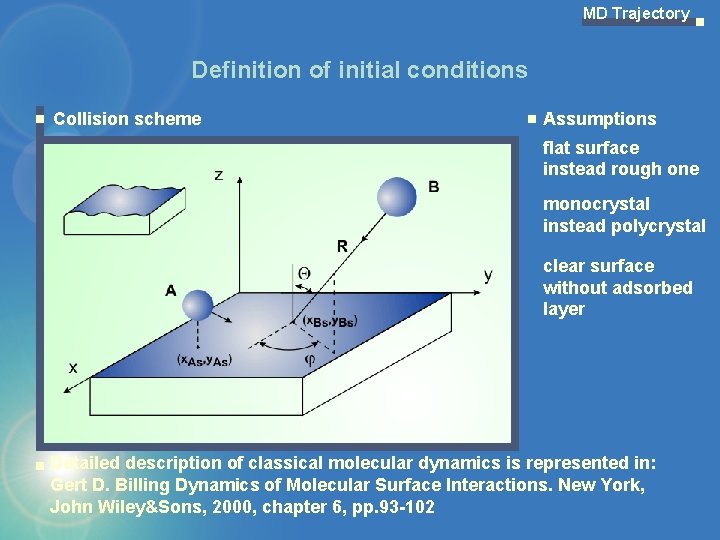
MD Trajectory Definition of initial conditions Collision scheme Assumptions flat surface instead rough one monocrystal instead polycrystal clear surface without adsorbed layer Detailed description of classical molecular dynamics is represented in: Gert D. Billing Dynamics of Molecular Surface Interactions. New York, John Wiley&Sons, 2000, chapter 6, pp. 93 -102

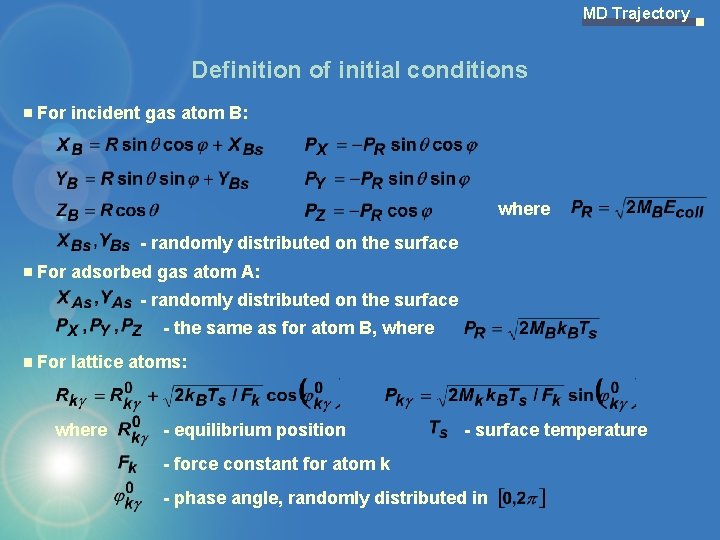
MD Trajectory Definition of initial conditions For incident gas atom B: where - randomly distributed on the surface For adsorbed gas atom A: - randomly distributed on the surface - the same as for atom B, where For lattice atoms: where - equilibrium position - surface temperature - force constant for atom k - phase angle, randomly distributed in

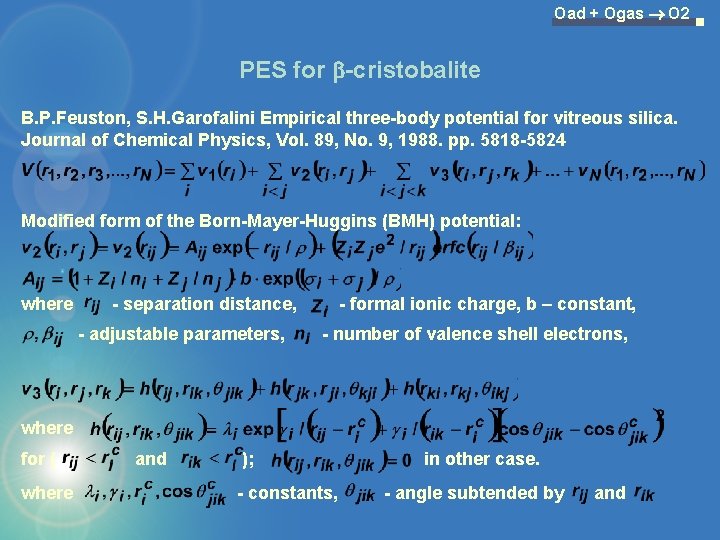
Oad + Ogas O 2 PES for b-cristobalite B. P. Feuston, S. H. Garofalini Empirical three-body potential for vitreous silica. Journal of Chemical Physics, Vol. 89, No. 9, 1988. pp. 5818 -5824 Modified form of the Born-Mayer-Huggins (BMH) potential: where - separation distance, - adjustable parameters, - formal ionic charge, b – constant, - number of valence shell electrons, where for ( where and ); - constants, in other case. - angle subtended by and

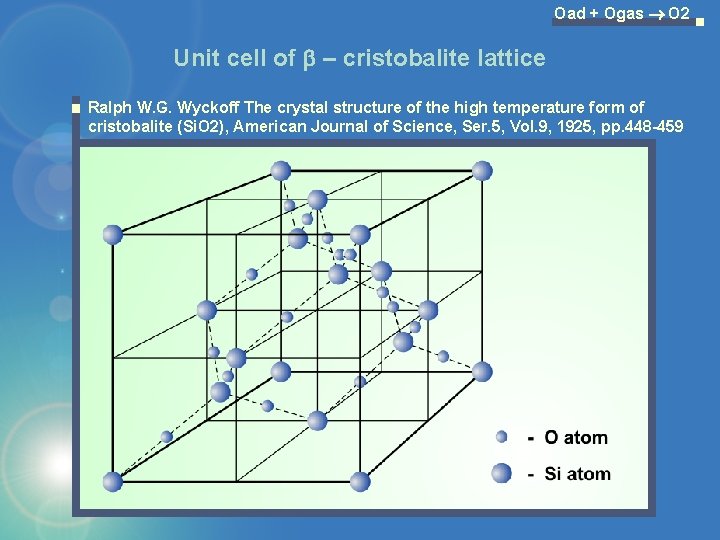
Oad + Ogas O 2 Unit cell of b – cristobalite lattice Ralph W. G. Wyckoff The crystal structure of the high temperature form of cristobalite (Si. O 2), American Journal of Science, Ser. 5, Vol. 9, 1925, pp. 448 -459

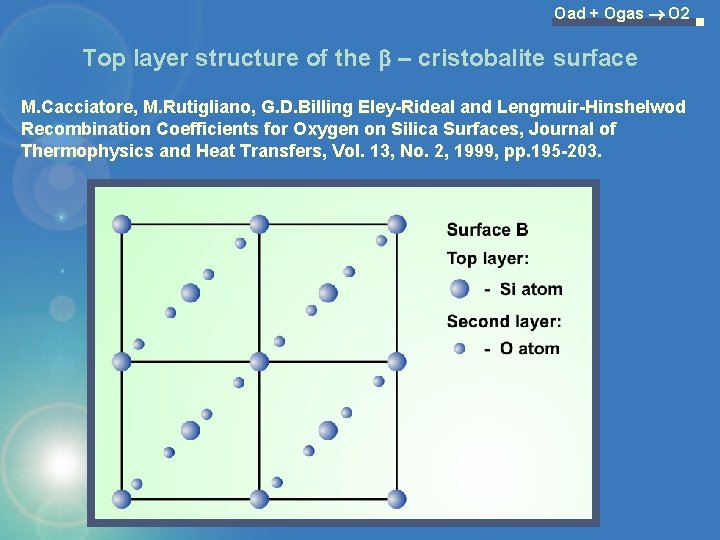
Oad + Ogas O 2 Top layer structure of the b – cristobalite surface M. Cacciatore, M. Rutigliano, G. D. Billing Eley-Rideal and Lengmuir-Hinshelwod Recombination Coefficients for Oxygen on Silica Surfaces, Journal of Thermophysics and Heat Transfers, Vol. 13, No. 2, 1999, pp. 195 -203.

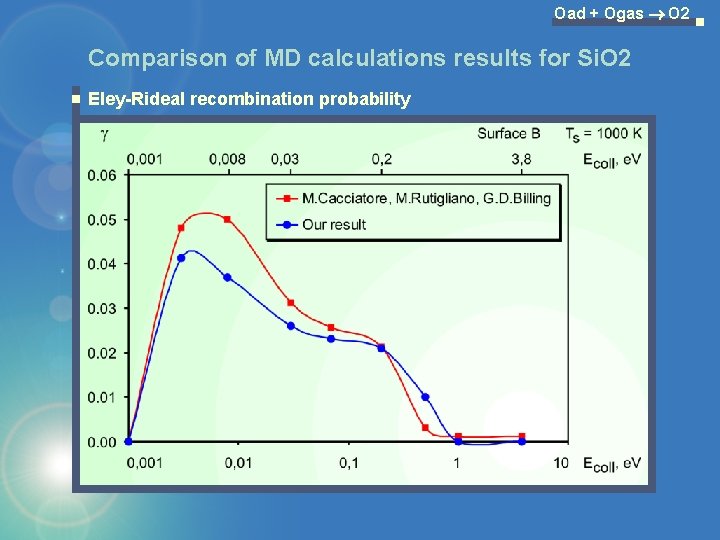
Oad + Ogas O 2 Comparison of MD calculations results for Si. O 2 Eley-Rideal recombination probability

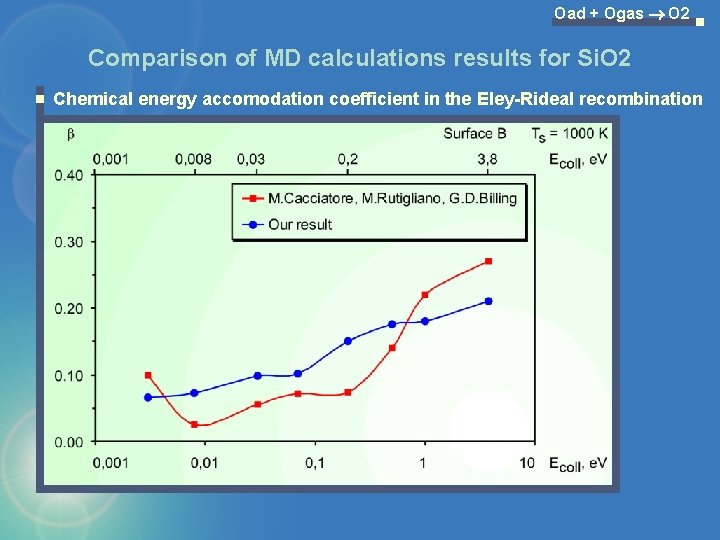
Oad + Ogas O 2 Comparison of MD calculations results for Si. O 2 Chemical energy accomodation coefficient in the Eley-Rideal recombination

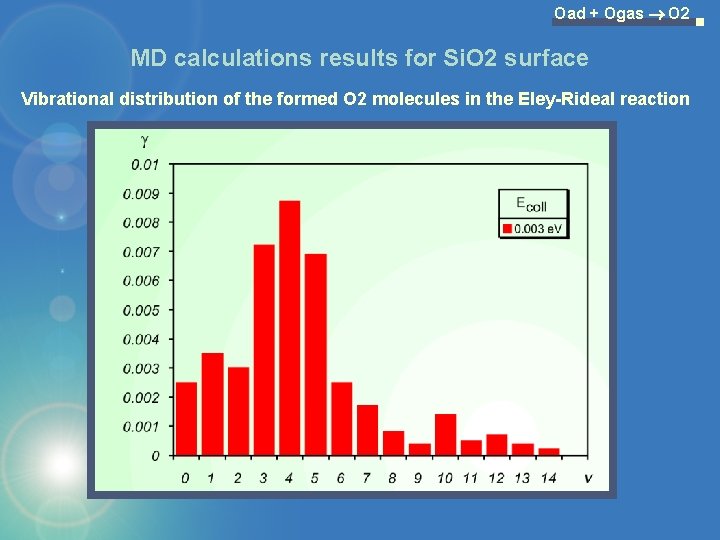
Oad + Ogas O 2 MD calculations results for Si. O 2 surface Vibrational distribution of the formed O 2 molecules in the Eley-Rideal reaction

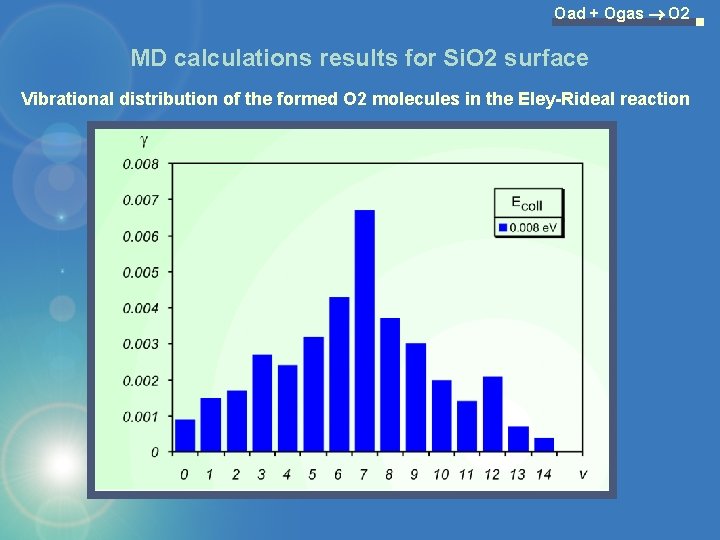
Oad + Ogas O 2 MD calculations results for Si. O 2 surface Vibrational distribution of the formed O 2 molecules in the Eley-Rideal reaction

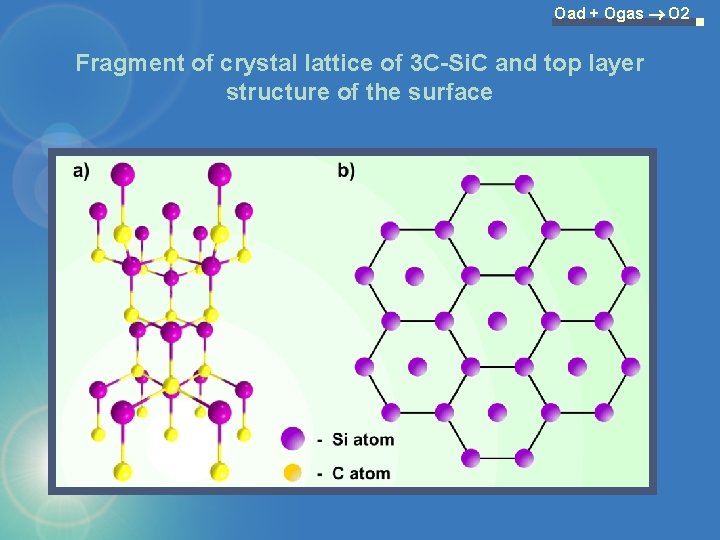
Oad + Ogas O 2 Fragment of crystal lattice of 3 C-Si. C and top layer structure of the surface

Oad + Ogas O 2 Comparison of MD calculations results Eley-Rideal recombination probability

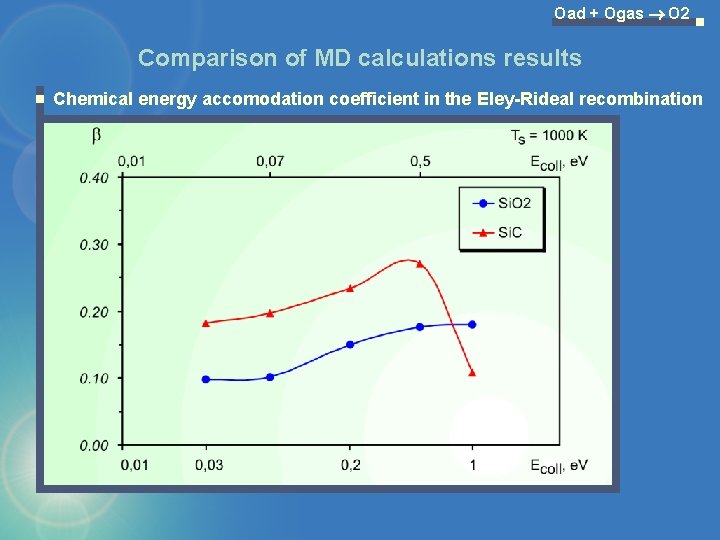
Oad + Ogas O 2 Comparison of MD calculations results Chemical energy accomodation coefficient in the Eley-Rideal recombination

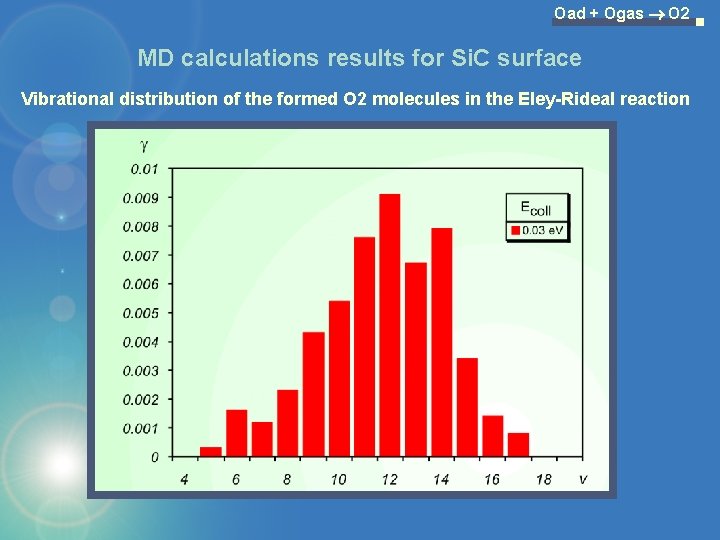
Oad + Ogas O 2 MD calculations results for Si. C surface Vibrational distribution of the formed O 2 molecules in the Eley-Rideal reaction

Oad + Ogas O 2 MD calculations results for Si. C surface Vibrational distribution of the formed O 2 molecules in the Eley-Rideal reaction

Thank you for your attention