Molecular Dynamics P OjedaMay B Bryds Y Li
Molecular Dynamics P. Ojeda-May, B. Brydsö, Y. Li, J. Eriksson

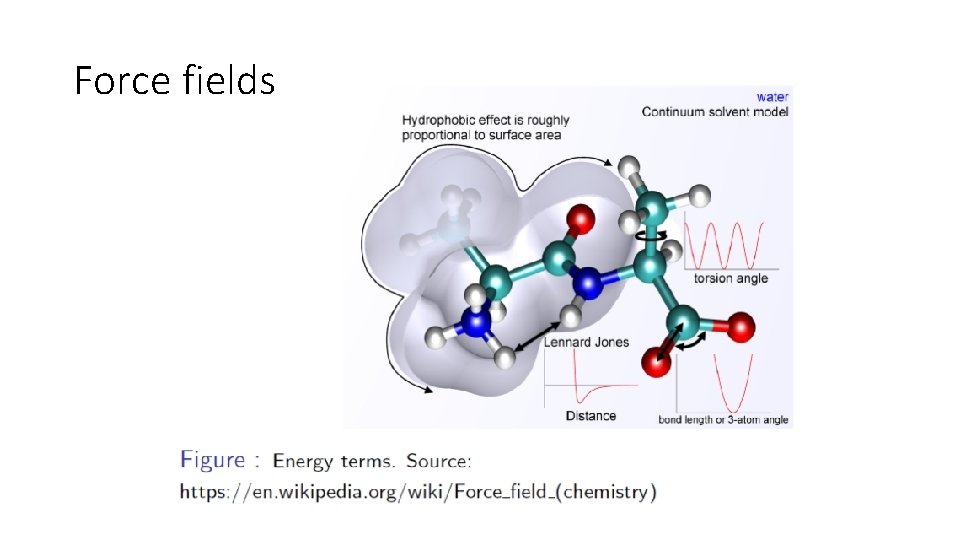
Force fields
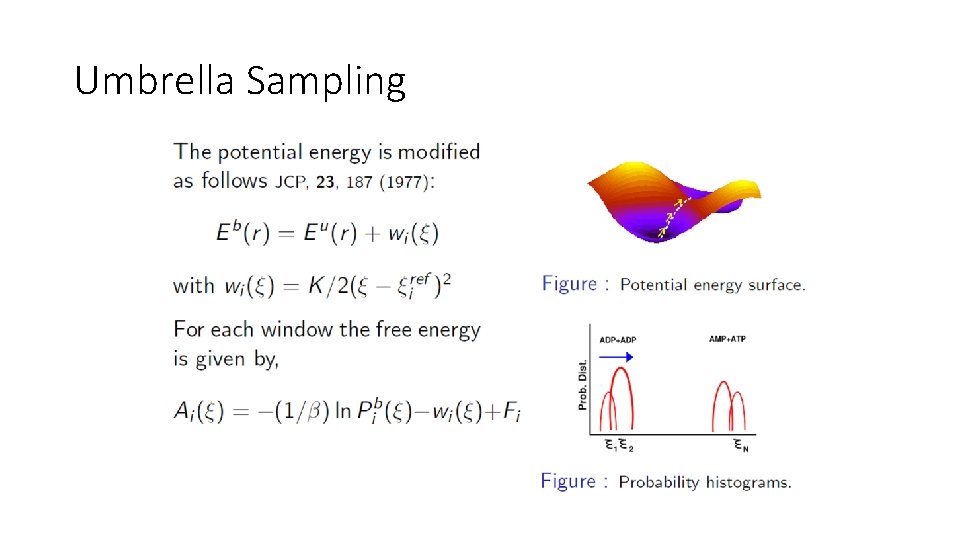
Umbrella Sampling

String method

Metadynamics P. Ojeda-May, B. Brydsoe, J. Eriksson

Error JCP B, 109, 6714 (2005).
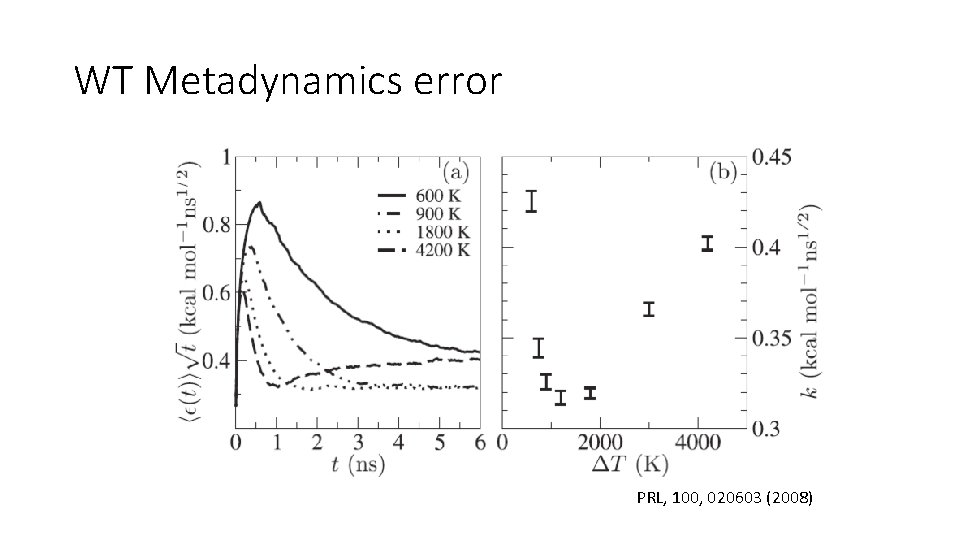
WT Metadynamics error PRL, 100, 020603 (2008)

Na. Cl system JCTC, 2018, Yi Yao and Yosuke Kanai
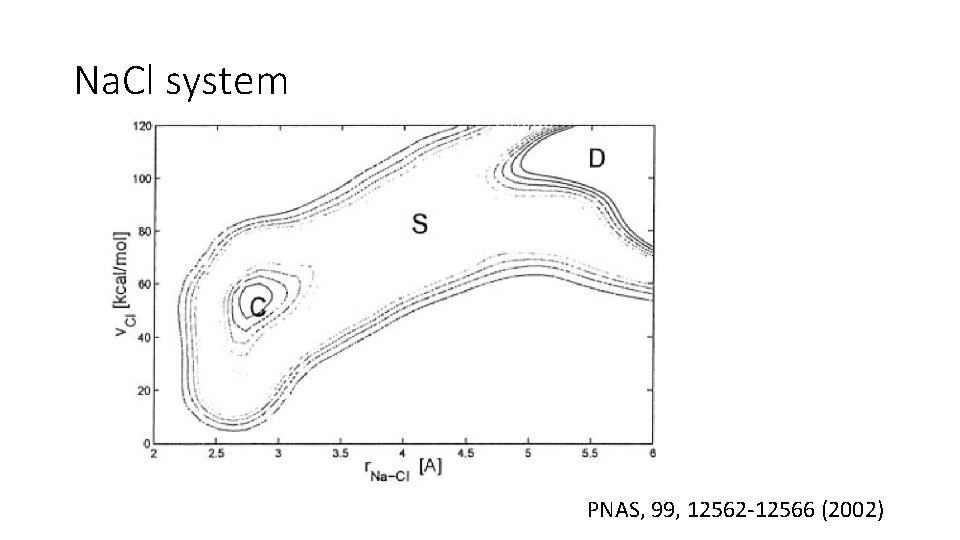
Na. Cl system PNAS, 99, 12562 -12566 (2002)
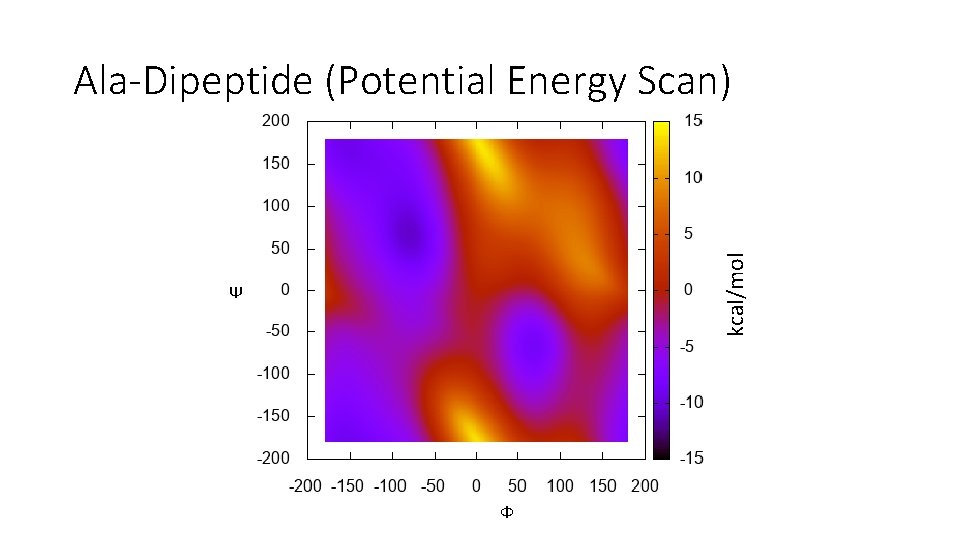
kcal/mol Ala-Dipeptide (Potential Energy Scan) Ψ Ф

Kinase system JACS, 131, 244 (2009)

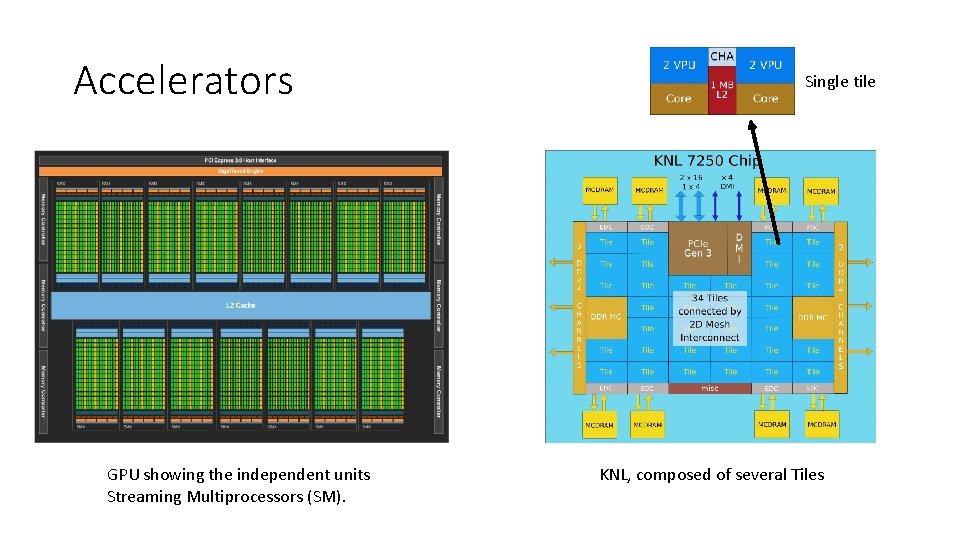
Accelerators GPU showing the independent units Streaming Multiprocessors (SM). Single tile KNL, composed of several Tiles


NAMD files • . pdb coordinates • Parameter file • X-plor topology file • . namd (input for the simulation)
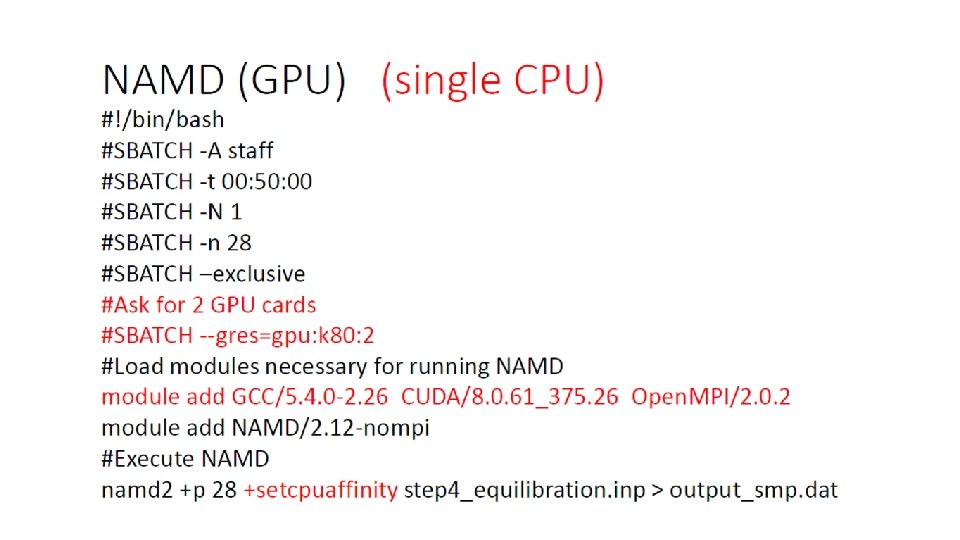

KNL Thread affinity #export OMP_PROC_BIND=spread, close, etc. #export OMP_PLACES=threads, cores, etc. export OMP_NUM_THREADS=4 srun -n 16 -c 4 --cpu_bind=cores. /xthi Hello from rank 0, thread 0, on b-cn 1209. hpc 2 n. umu. se. (core affinity = 0) Hello from rank 0, thread 1, on b-cn 1209. hpc 2 n. umu. se. (core affinity = 68) Hello from rank 0, thread 2, on b-cn 1209. hpc 2 n. umu. se. (core affinity = 136) Hello from rank 0, thread 3, on b-cn 1209. hpc 2 n. umu. se. (core affinity = 204) Hello from rank 1, thread 0, on b-cn 1209. hpc 2 n. umu. se. (core affinity = 1) Hello from rank 1, thread 1, on b-cn 1209. hpc 2 n. umu. se. (core affinity = 69) Hello from rank 1, thread 2, on b-cn 1209. hpc 2 n. umu. se. (core affinity = 137) Hello from rank 1, thread 3, on b-cn 1209. hpc 2 n. umu. se. (core affinity = 205)
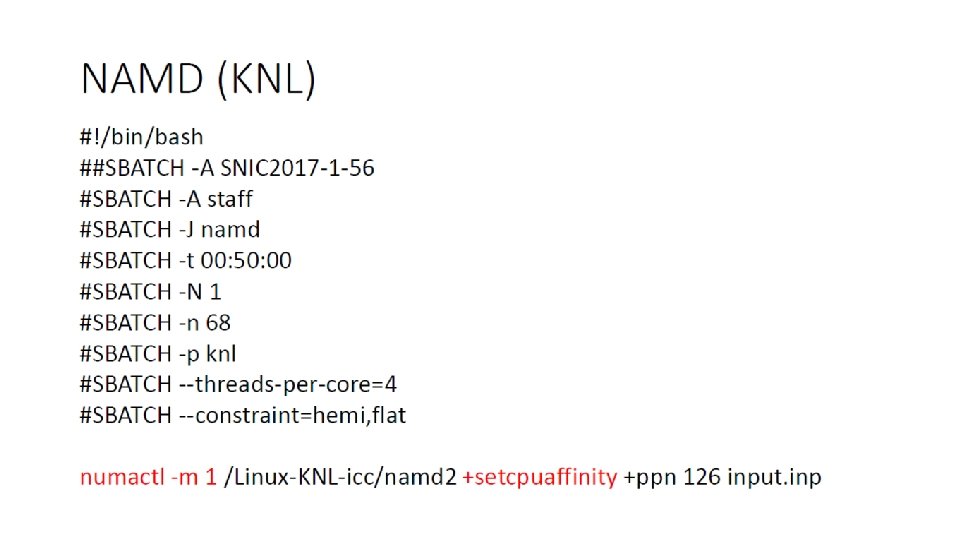
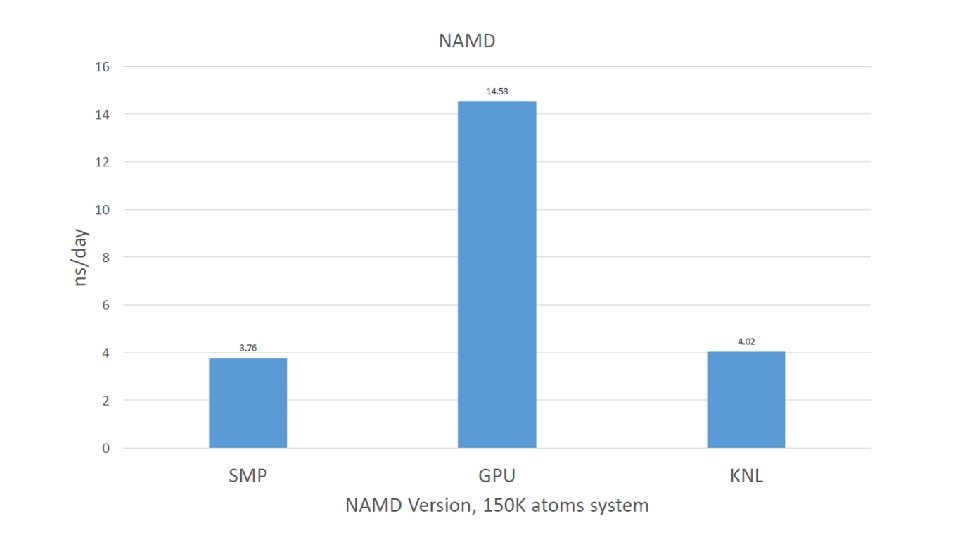

Accelerated MD Yaozong Li, P. Ojeda-May
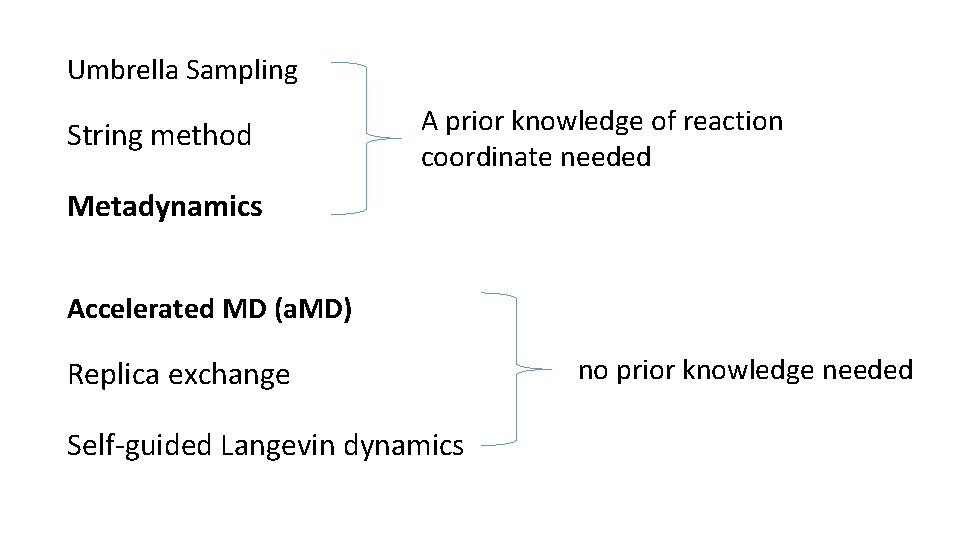
Umbrella Sampling String method A prior knowledge of reaction coordinate needed Metadynamics Accelerated MD (a. MD) Replica exchange Self-guided Langevin dynamics no prior knowledge needed

Why a. MD Fastly observe rare events that conventional MD cannot sample in a feasible simulation time. no reaction coordinate needs to be defined a priori.
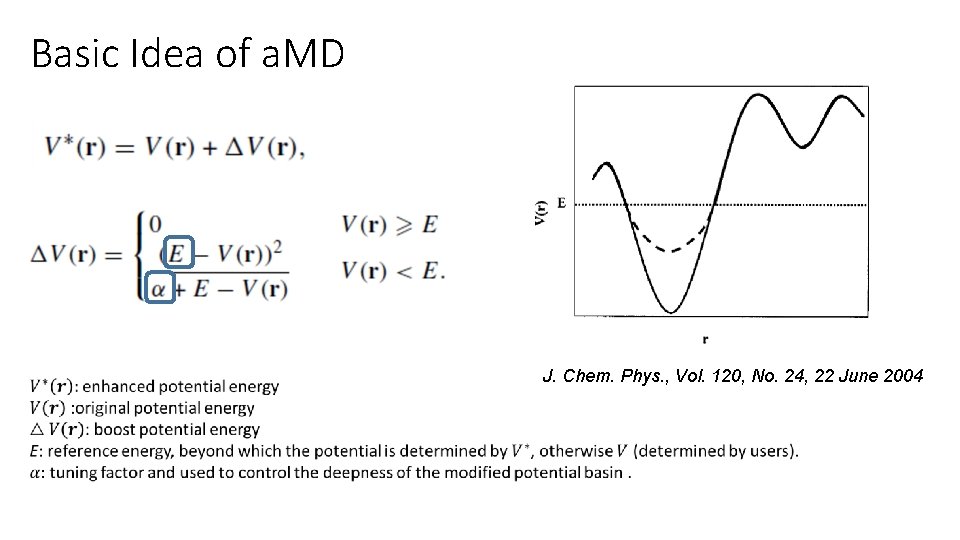
Basic Idea of a. MD J. Chem. Phys. , Vol. 120, No. 24, 22 June 2004
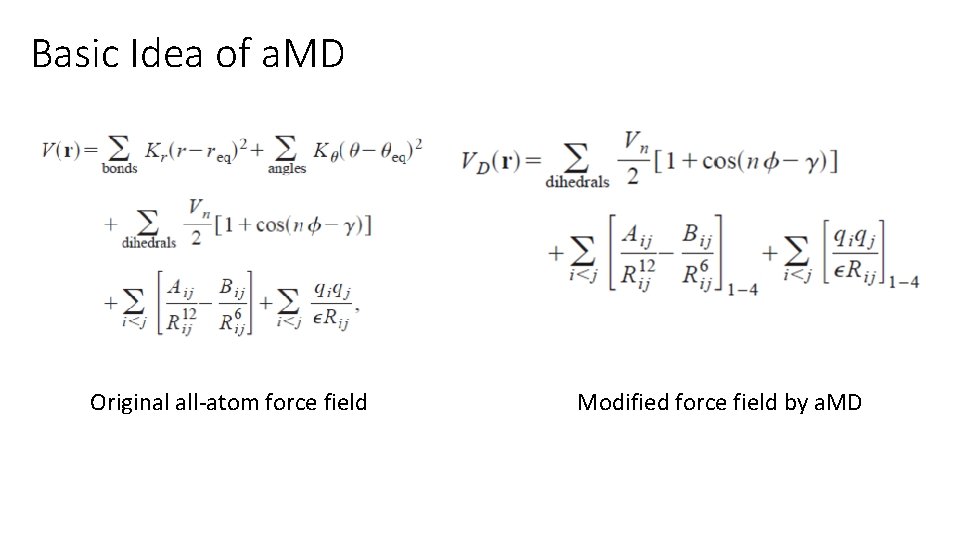
Basic Idea of a. MD Original all-atom force field Modified force field by a. MD
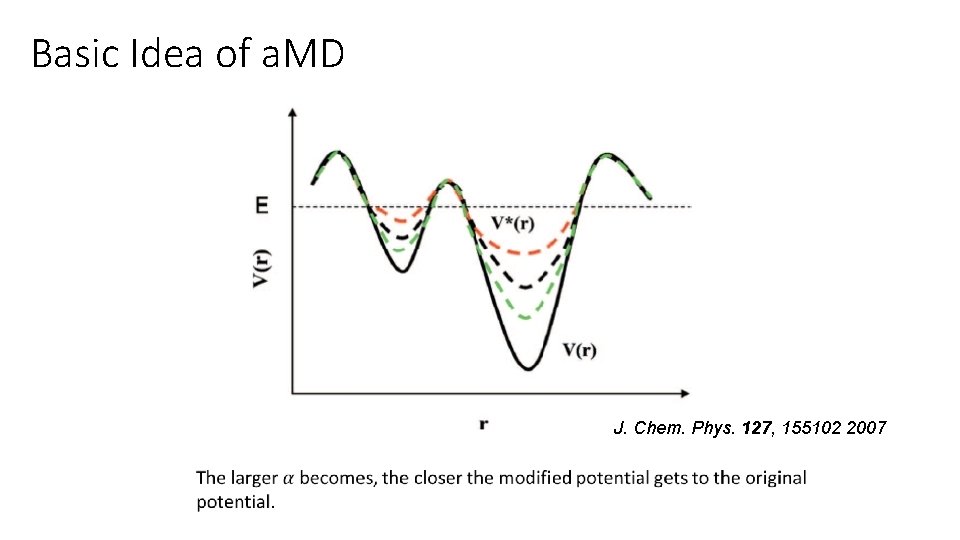
Basic Idea of a. MD J. Chem. Phys. 127, 155102 2007
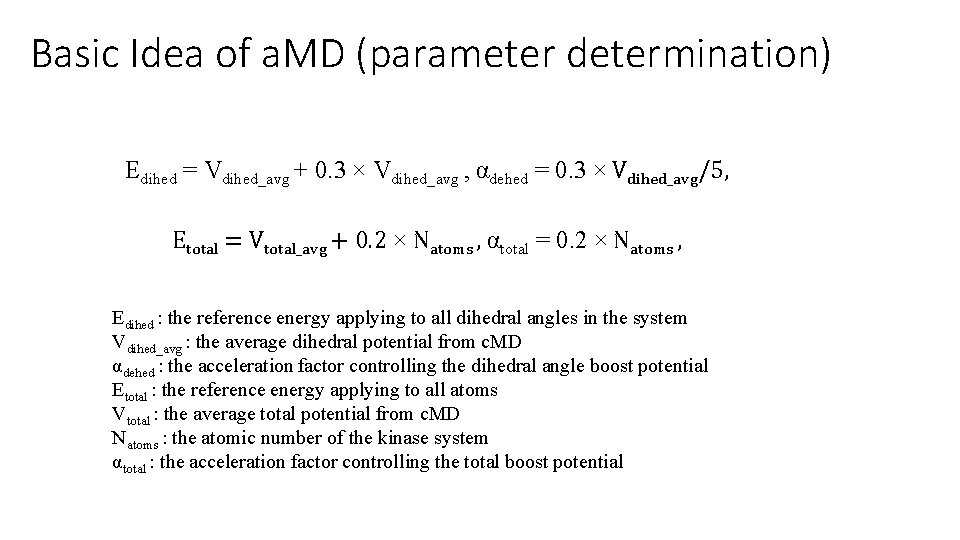
Basic Idea of a. MD (parameter determination) Edihed = Vdihed_avg + 0. 3 × Vdihed_avg , αdehed = 0. 3 × Vdihed_avg/5, Etotal = Vtotal_avg + 0. 2 × Natoms , αtotal = 0. 2 × Natoms , Edihed : the reference energy applying to all dihedral angles in the system Vdihed_avg : the average dihedral potential from c. MD αdehed : the acceleration factor controlling the dihedral angle boost potential Etotal : the reference energy applying to all atoms Vtotal : the average total potential from c. MD Natoms : the atomic number of the kinase system αtotal : the acceleration factor controlling the total boost potential
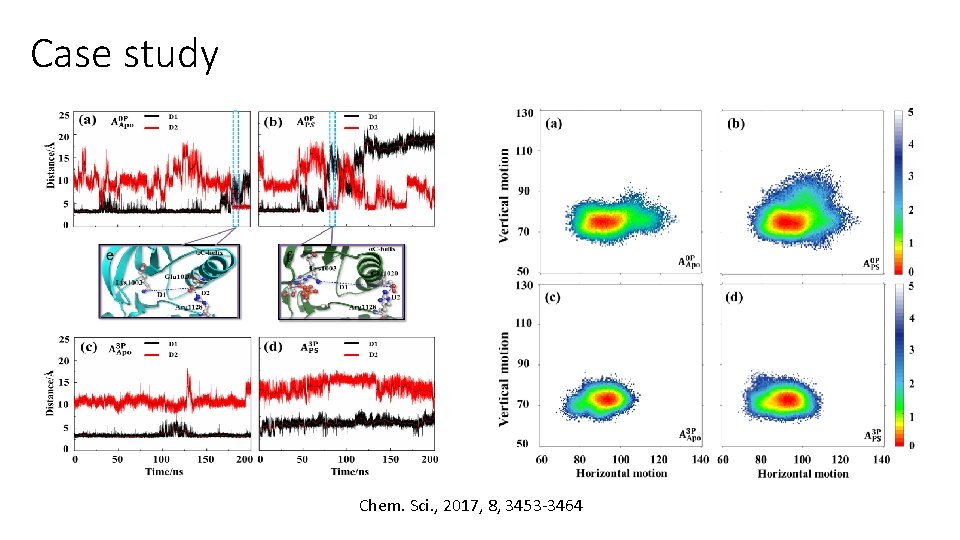
Case study Chem. Sci. , 2017, 8, 3453 -3464
- Slides: 28