Molecular Clock Lineagespecific evolutionary rate Xuhua Xia xxiauottawa





- Slides: 23

Molecular Clock: Lineage-specific evolutionary rate Xuhua Xia xxia@uottawa. ca http: //dambe. bio. uottawa. ca

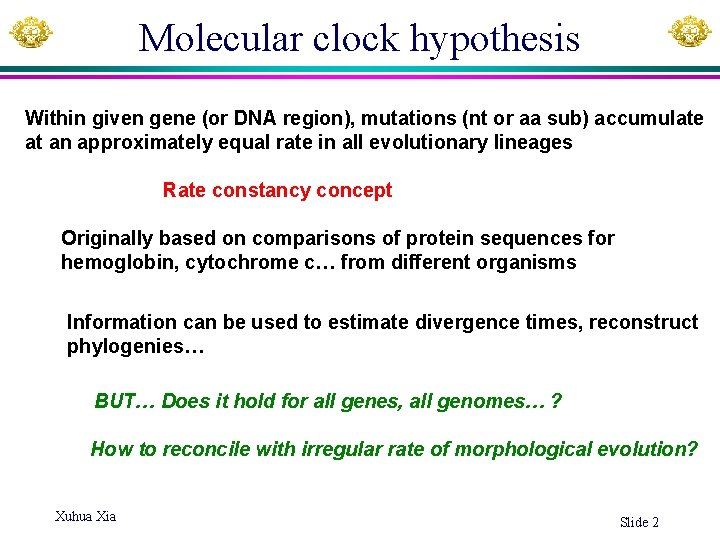
Molecular clock hypothesis Within given gene (or DNA region), mutations (nt or aa sub) accumulate at an approximately equal rate in all evolutionary lineages Rate constancy concept Originally based on comparisons of protein sequences for hemoglobin, cytochrome c… from different organisms Information can be used to estimate divergence times, reconstruct phylogenies… BUT… Does it hold for all genes, all genomes… ? How to reconcile with irregular rate of morphological evolution? Xuhua Xia Slide 2

Clock-like substitution rate Fig. 4. 15 Roughly linear relationship Slope: evolutionary rate in subs/site/my Combined data for hemoglobins, cytochrome c & fibrinopeptide several apparent deviations: slowdown in rate after divergence of ape/monkey lineages & acceleration after split of horse/donkey lineages What would figure look like without combining genes

Different rates among genes Linear relationship between # substitutions and geological time Rates of aa substitution vary among proteins Same intercept but different slopes Fibrinopeptides have a much higher proportion of neutral mutations because these peptides are cut of fibrinogen to form fibrin to activate the blood-clotting reaction (i. e, . they are not functionally constrained) Selection by war and peace: clotters and bleeders Xuhua Xia Alberts Fig. 5. 1 Slide 4

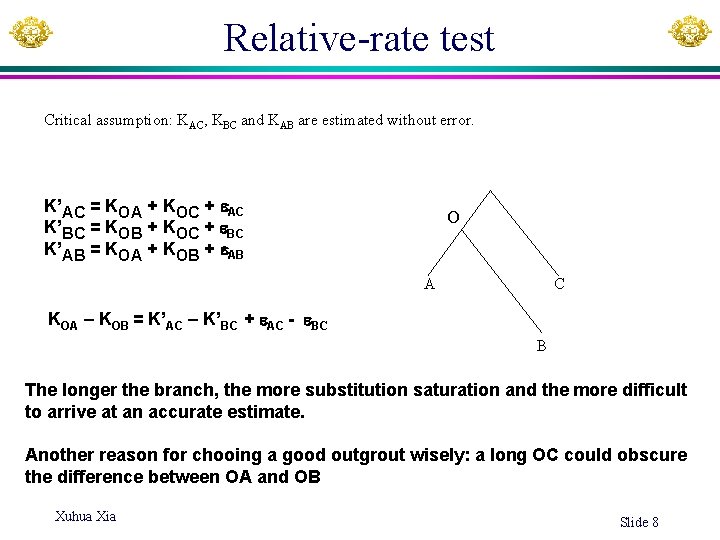
Relative-rate test To compare rates in lineages A and B, use C as reference species If constant rate (clock hypothesis H 0), then H 0: KOA = KOB KAC = KOA + KOC (1) so d = KOA – KOB = 0 KBC = KOB + KOC (2) How to get d? KAB = KOA + KOB (3) So O KOA = (KAC + KAB - KBC ) / 2 KOB = (KAB + KBC - KAC ) / 2 KOC = (KAC + KBC - KAB ) / 2 A B Xuhua Xia C d = KOA – KOB = KAC – KBC The importance of chooing a good outgroup: a long OC could obscure the difference between OA and OB

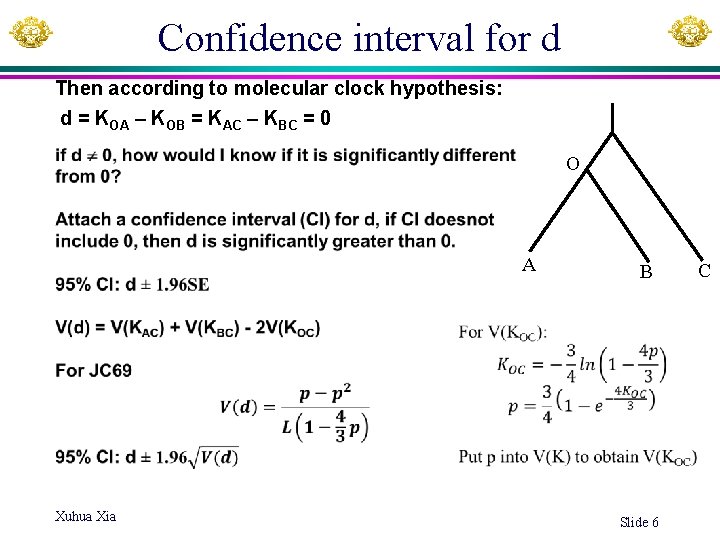
Confidence interval for d Then according to molecular clock hypothesis: d = KOA – KOB = KAC – KBC = 0 O A B Xuhua Xia Slide 6 C

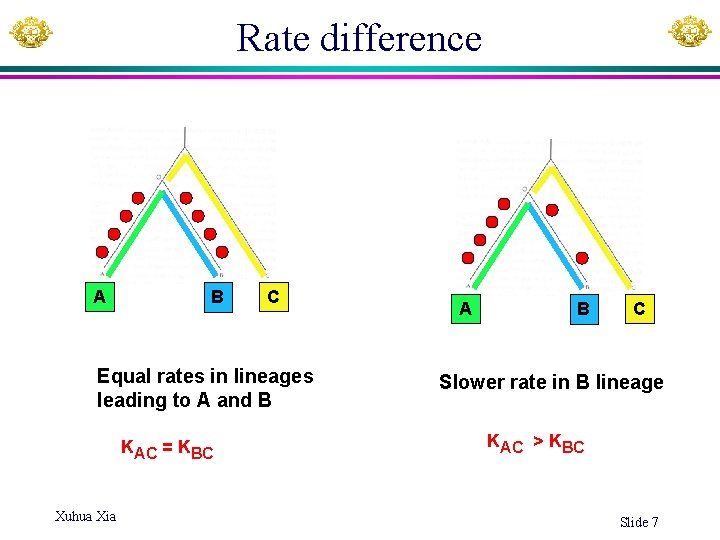
Rate difference A B C Equal rates in lineages leading to A and B KAC = KBC Xuhua Xia A B C Slower rate in B lineage KAC > KBC Slide 7

Relative-rate test Critical assumption: KAC, KBC and KAB are estimated without error. K’AC = KOA + KOC + AC K’BC = KOB + KOC + BC K’AB = KOA + KOB + AB O A C KOA – KOB = K’AC – K’BC + AC - BC B The longer the branch, the more substitution saturation and the more difficult to arrive at an accurate estimate. Another reason for chooing a good outgrout wisely: a long OC could obscure the difference between OA and OB Xuhua Xia Slide 8

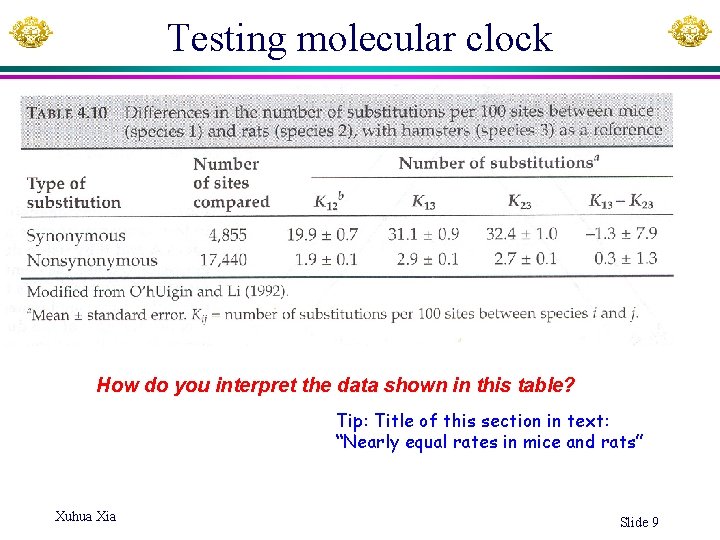
Testing molecular clock How do you interpret the data shown in this table? Tip: Title of this section in text: “Nearly equal rates in mice and rats” Xuhua Xia Slide 9

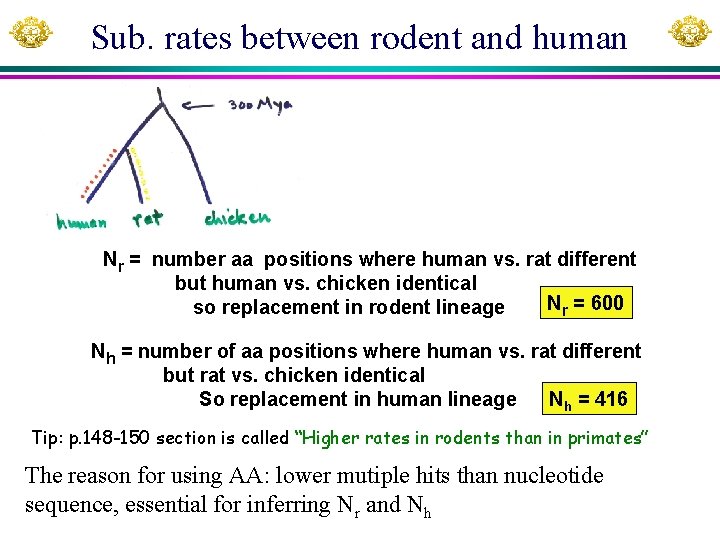
Sub. rates between rodent and human Nr = number aa positions where human vs. rat different but human vs. chicken identical Nr = 600 so replacement in rodent lineage Nh = number of aa positions where human vs. rat different but rat vs. chicken identical So replacement in human lineage Nh = 416 Tip: p. 148 -150 section is called “Higher rates in rodents than in primates” The reason for using AA: lower mutiple hits than nucleotide sequence, essential for inferring Nr and Nh

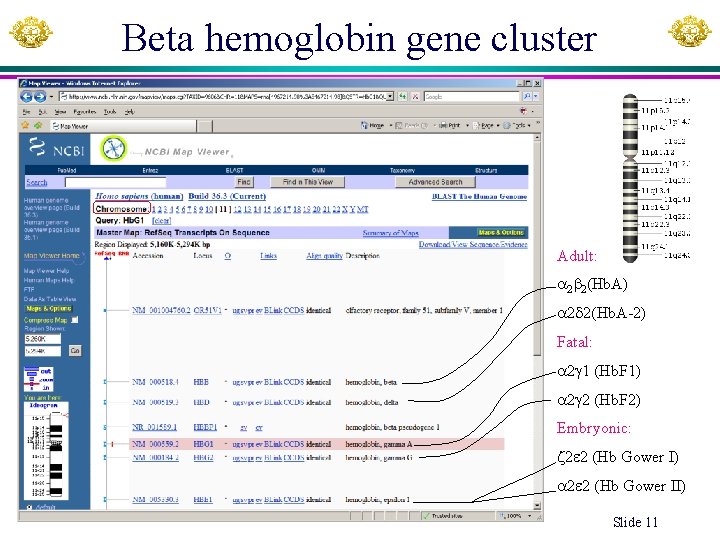
Beta hemoglobin gene cluster Adult: 2 2(Hb. A) 2 2(Hb. A-2) Fatal: 2 1 (Hb. F 1) 2 2 (Hb. F 2) Embryonic: 2 2 (Hb Gower I) 2 2 (Hb Gower II) Xuhua Xia Slide 11

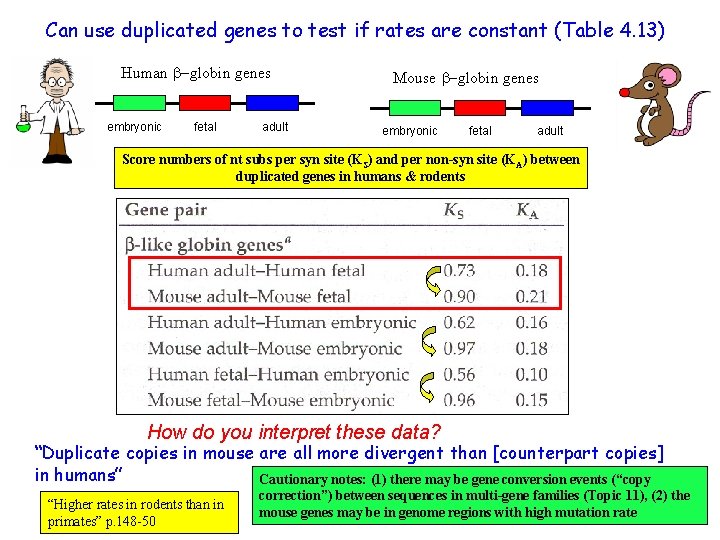
Can use duplicated genes to test if rates are constant (Table 4. 13) Human -globin genes embryonic fetal adult Mouse -globin genes embryonic fetal adult Score numbers of nt subs per syn site (KS) and per non-syn site (KA) between duplicated genes in humans & rodents How do you interpret these data? “Duplicate copies in mouse are all more divergent than [counterpart copies] in humans” Cautionary notes: (1) there may be gene conversion events (“copy “Higher rates in rodents than in Xuhua Xia primates” p. 148 -50 correction”) between sequences in multi-gene families (Topic 11), (2) the mouse genes may be in genome regions with high mutation rate

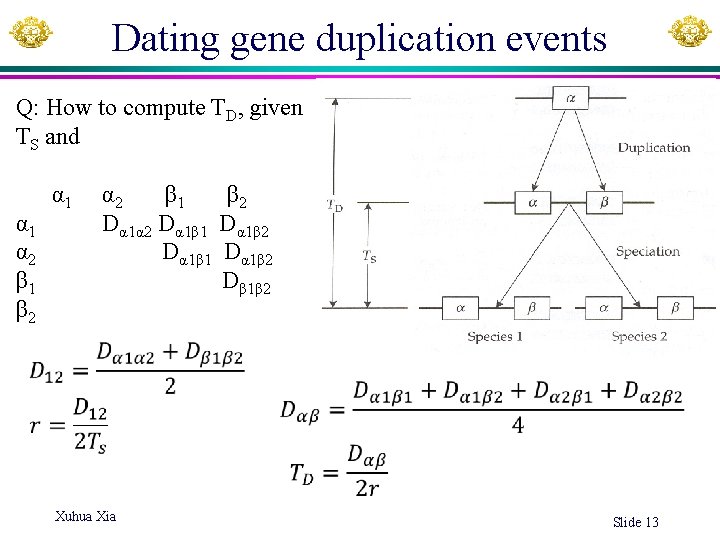
Dating gene duplication events Q: How to compute TD, given TS and α 1 α 2 β 1 β 2 α 1 Dα 1α 2 Dα 1β 1 Dα 1β 2 α 2 Dα 1β 1 Dα 1β 2 β 1 Dβ 1β 2 Xuhua Xia Slide 13

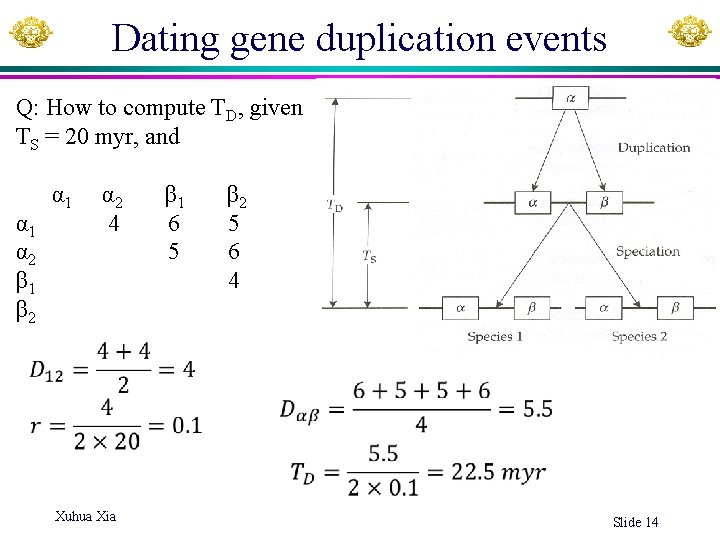
Dating gene duplication events Q: How to compute TD, given TS = 20 myr, and α 1 α 2 β 1 β 2 α 1 4 6 5 α 2 5 6 β 1 4 β 2 Xuhua Xia Slide 14

Causes of rate differences (p. 152) • Mutation rates – Generation time – Metabolic rate (e. g. , high aerobic respiration leads to mutagenic effects of oxygen free radicals) – DNA repair – Gene conversion in multi-gene families • Substitution rates: – population size – genetic variation – purifying selection or positive selection • Different genomic background (known different rates at different genomic regions) Xuhua Xia Slide 15

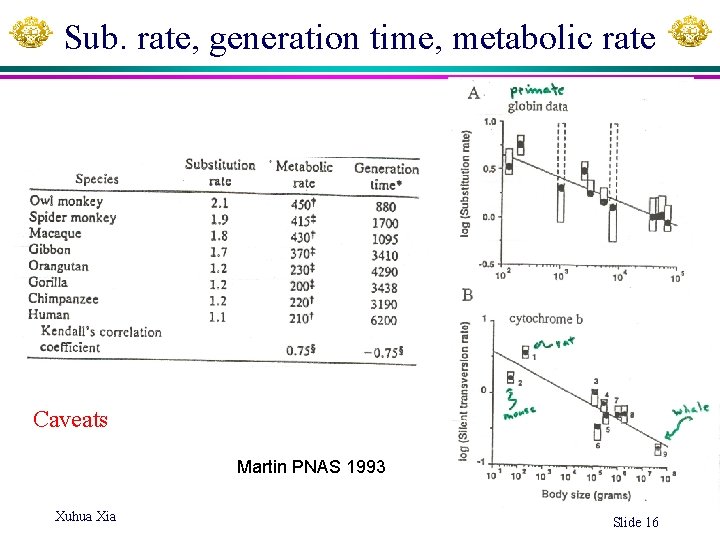
Sub. rate, generation time, metabolic rate Caveats Martin PNAS 1993 Xuhua Xia Slide 16

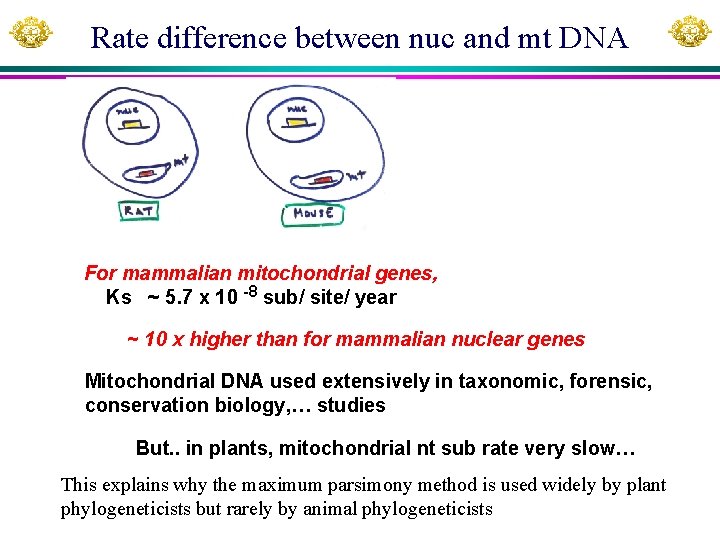
Rate difference between nuc and mt DNA For mammalian mitochondrial genes, Ks ~ 5. 7 x 10 -8 sub/ site/ year ~ 10 x higher than for mammalian nuclear genes Mitochondrial DNA used extensively in taxonomic, forensic, conservation biology, … studies But. . in plants, mitochondrial nt sub rate very slow… This explains why the maximum parsimony method is used widely by plant phylogeneticists but rarely by animal phylogeneticists

Relative rates for nuclear, chloroplast and mitochondrial genes in plant cells? In plants, mitochondrial rates of nt sub are much slower than nuclear (or chloroplast) … whereas mammalian mito rate ~ 10 x faster than nuclear L = # sites

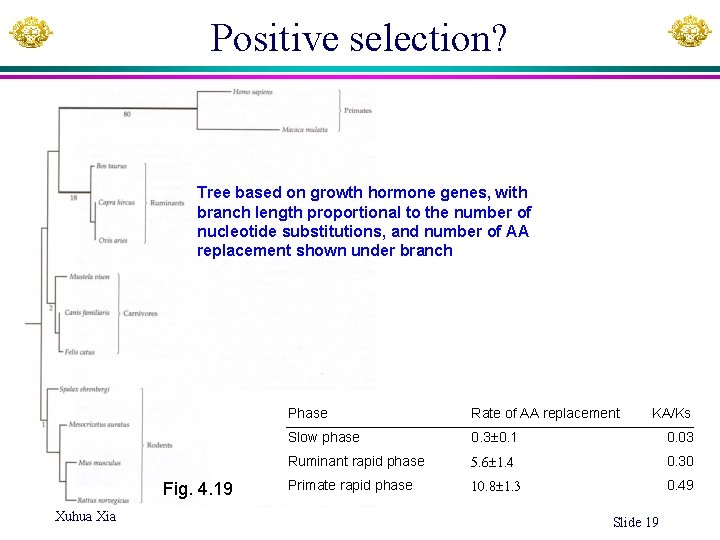
Positive selection? Tree based on growth hormone genes, with branch length proportional to the number of nucleotide substitutions, and number of AA replacement shown under branch Fig. 4. 19 Xuhua Xia Phase Rate of AA replacement KA/Ks Slow phase 0. 3± 0. 1 0. 03 Ruminant rapid phase 5. 6± 1. 4 0. 30 Primate rapid phase 10. 8± 1. 3 0. 49 Slide 19

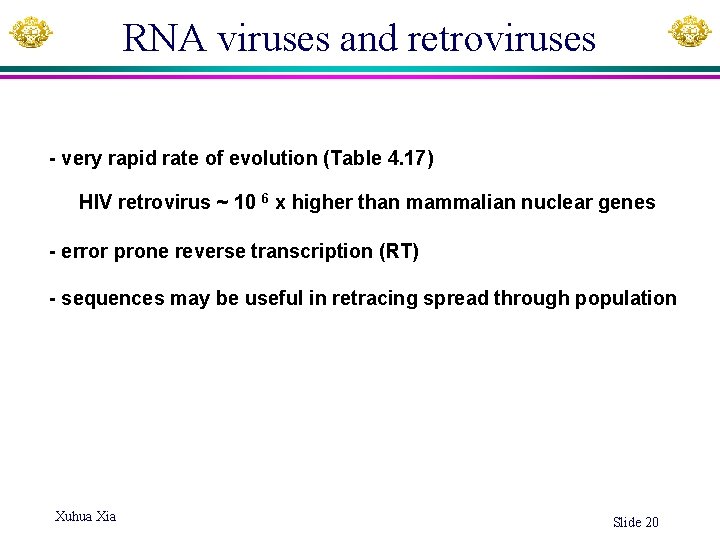
RNA viruses and retroviruses - very rapid rate of evolution (Table 4. 17) HIV retrovirus ~ 10 6 x higher than mammalian nuclear genes - error prone reverse transcription (RT) - sequences may be useful in retracing spread through population Xuhua Xia Slide 20

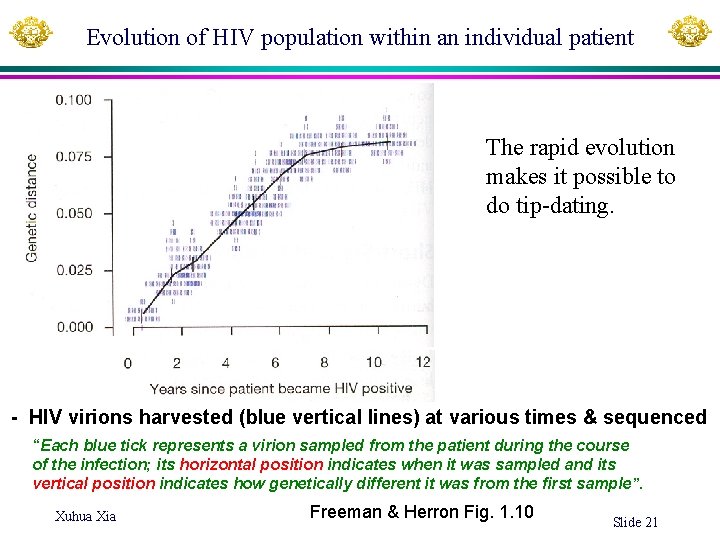
Evolution of HIV population within an individual patient The rapid evolution makes it possible to do tip-dating. - HIV virions harvested (blue vertical lines) at various times & sequenced “Each blue tick represents a virion sampled from the patient during the course of the infection; its horizontal position indicates when it was sampled and its vertical position indicates how genetically different it was from the first sample”. Xuhua Xia Freeman & Herron Fig. 1. 10 Slide 21

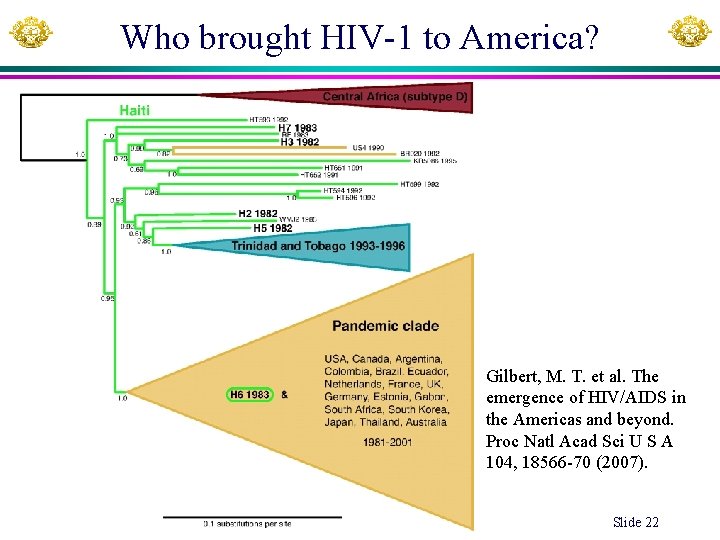
Who brought HIV-1 to America? Gilbert, M. T. et al. The emergence of HIV/AIDS in the Americas and beyond. Proc Natl Acad Sci U S A 104, 18566 -70 (2007). Xuhua Xia Slide 22

Who brought HIV-1 to America? Gilbert, M. T. et al. The emergence of HIV/AIDS in the Americas and beyond. Proc Natl Xuhua Xia Slide 23 Acad Sci U S A 104, 18566 -70 (2007).
 Bps courses uottawa
Bps courses uottawa Xuhua xia rate my prof
Xuhua xia rate my prof Xuhua xia
Xuhua xia Mega molecular
Mega molecular Arvore filogenetica
Arvore filogenetica Fast clock to slow clock synchronization
Fast clock to slow clock synchronization 1hr = 60 minutes
1hr = 60 minutes Dicots
Dicots Melting and boiling point of oxygen
Melting and boiling point of oxygen Ionic covalent metallic
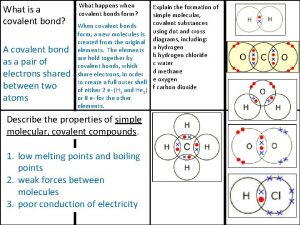
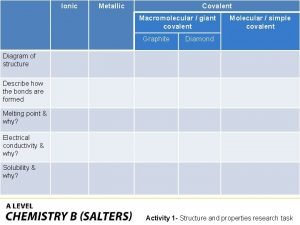
Ionic covalent metallic Giant molecular structure vs simple molecular structure
Giant molecular structure vs simple molecular structure Yuni xia
Yuni xia Lingxiao xia
Lingxiao xia Lirong xia
Lirong xia Guoxing xia
Guoxing xia A xia
A xia Xia dynasty government
Xia dynasty government Perfume xia xiang
Perfume xia xiang Guoxing xia
Guoxing xia Swbat
Swbat Lirong xia rpi
Lirong xia rpi Jennifer xia
Jennifer xia Derek xia
Derek xia Albert xia
Albert xia